Professional Documents
Culture Documents
Fusion Division Fusion Division
Fusion Division Fusion Division
Uploaded by
Giuliano CiolacuOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Fusion Division Fusion Division
Fusion Division Fusion Division
Uploaded by
Giuliano CiolacuCopyright:
Available Formats
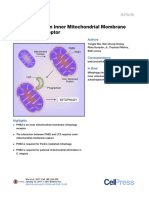
SnapShot: Mitochondrial Dynamics
Yasushi Tamura,1 Kie Itoh,1 and Hiromi Sesaki,1
1
Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Mitochondrialfusion-divisioncycle
Mitochondrialshape
Fusion < Division
Drp1/Dnm1
Fusion Division
Fusion > Division
Tubular
Elongated
Fragmented
Drp1/Dnm1 receptor
Mfn1 and 2/Fzo1
Opa1/Mgm1
MEF
Yeast
10 m
Fragmented with
swollen cristae
5 m
Enlarged
Fusionmachinery
Mammals
CYTOSOL
Positive regulator
for Mfn2
Bax
Bcl-xL
Ubiquitin ligase for Mfn1 and 2
Parkin Pink1 MITOL/March5
Yeast
CYTOSOL
Mfn1
Mfn2
Ub
Proteasome
degraded
Ub
Negative regulator
for Mfn1 and 2
MIB
SCF Mdm30
OM
Ugo1
Fzo1
OM
IMS
Ubiquitin ligase
for Fzo1
i-AAA
s-Opa1
IMS
s-Mgm1
IM
m-AAA
Parl
Oma1
IM
l-Opa1
l-Mgm1
Pcp1
Processing peptidases
for Opa1
Processing peptidases
for Mgm1
HUMAN DISEASE ASSOCIATION
Mfn2: Charcot-Marie-Tooth type 2A
Parkin and Pink1: Parkinsons disease
Opa1: Autosomal dominant optic atrophy type 1
MATRIX
MATRIX
Divisionmachinery
Mammals
Yeast
Posttranslational
modulators for Drp1
CYTOSOL
Algae
CYTOSOL
CYTOSOL
PKA
CaMKla P Calcineurin
Cdk1-cyclin B
NO
Senp5
SUMO
MAPL
Ub
G
Drp1
IM
Dnm1
Dnm1
Dnm1
G
Num1
Fis1
Drp1 receptors
Mff
MiD49/MiD51
Fis1
Mda1
Fis1
OM
IMS
IMS
IM
IM
ZED
MATRIX
HUMAN DISEASE ASSOCIATION
Drp1: Postneonatal death with neurodevelopmental disorders
GDAP1: Charcot-Marie-Tooth type 4A
MATRIX
1158 Cell145,June24,20112011ElsevierInc. DOI10.1016/j.cell.2011.06.018
G
Dnm1
Dnm1
Mdm36
OM
GDAP1
Cortical actin
Mdv1
Caf4
MITOL/March5
OM
IMS
FtsZ
MATRIX
Seeonlineversionforlegendandreferences.
SnapShot: Mitochondrial Dynamics
Yasushi Tamura,1 Kie Itoh,1 and Hiromi Sesaki,1
1
Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
Mitochondrial Fusion and Division
Mitochondria are tubular, highly dynamic organelles that continuously fuse and divide in a regulated manner. A balance of fusion and division controls mitochondrial morphology; imbalanced dynamics leads to altered morphology, which is associated with a variety of pathological conditions. When fusion is decreased, mitochondria fragment into
small, spherical mitochondria that are often characterized by swollen cristae and impaired respiratory functions. When division is inhibited, tubular mitochondria fuse, generating
elongated mitochondrial tubules with increased connectivity. In some neurons, however, decreased division leads to enlarged, spherical mitochondria.
Highlighting the importance of mitochondrial fusion and division in human health and disease, mutations in mitochondrial dynamics components have recently been linked
to several neurodevelopmental and neurodegenerative diseases, including a birth defect with multiple neurological disorders (Drp1), Parkinsons disease (Parkin and Pink1),
autosomal dominant optic atrophy type 1 (Opa1), and Charcot-Marie-Tooth neuropathies (Mfn2 and GDAP1). In addition, although Alzheimers and Huntingtons diseases are not
associated with such mutations, they do show altered activity and abundance of mitochondrial dynamics components.
Core Machineries for Mitochondrial Fusion and Division
Dynamin-related GTPases feature prominently in mitochondrial fusion and division. Complexes of mitofusin 1 and 2 (Fzo1) control outer-membrane fusion, whereas Opa1 (Mgm1)
mediates inner-membrane fusion. In addition to its role in fusion, Opa1 (Mgm1) has been implicated in direct control of cristae junctions. For mitochondrial division, Drp1 (Dnm1)
is recruited to the organelle surface, where it assembles into spiral filaments that are thought to generate mechanical force, constricting and pinching off the mitochondria.
Regulation of Fusion
Although outer- and inner-membrane fusion events are coordinated, these processes require separate machineries. In the outer membrane, levels of mitofusins (Fzo1), which are
regulated by ubiquitin proteasome pathway, influence the amount of organelle fusion. In mammals, mitofusins are ubiquitinated by two E3 ligases, MITOL/March5 and Parkin.
MITOL/March5 is located in the outer membrane and ubiquitinates mitofusin 1. Parkin is translocated to dysfunctional, depolarized mitochondria by the Pink1 kinase. Ubiquitination and proteasomal degradation of mitofusins inhibit refusion of damaged mitochondria and promote autophagic degradation of mitochondria. The ubiquitin proteasome
pathway plays a similar role in yeast, with the ubiquitin ligase SCFMdm30 regulating the levels of Fzo1.
Mitochondrial fusion can be modulated independently of the amount of fusion proteins. For instance, the proapoptotic Bcl-2 family members Bax and Bcl-xL stimulate mitofusin 2 activity, whereas a mitofusin-binding protein (MIB) inhibits mitofusin 1 and 2.
In the inner membrane, Opa1 (Mgm1) exists in two forms: l-Opa1 (l-Mgm1) is integral to the inner membrane, and s-Opa1 (s-Mgm1) is soluble as a result of proteolytic cleavage
of the integrated membrane form. Both are required for mitochondrial fusion. In mammals, several inner membrane-localized proteases cleave Opa1, including PARL, i-AAA,
m-AAA, and Oma1. In yeast, a homolog of PARL, Pcp1, cleaves Mgm1. Changes in matrix ATP levels and the membrane potential across the inner membrane affect Opa1 (Mgm1)
processing.
Because mitochondrial have two membranes, efficient fusion of the organelle requires coordination of outer- and inner-membrane fusion. Ugo1, an outer-membrane protein
binds Fzo1 and Mgm1, linking these two fusion events in yeast. The mammalian homolog of Ugo1 remains unidentified.
Regulation of Division Machinery
Multiple integral outer-membrane proteins recruit Drp1 (Dnm1) to mitochondria. In mammals, Mff and MiD49/51 interact directly with Drp1, anchoring it to the mitochondrial surface. Fis1 was the first outer-membrane protein identified as a tether for Drp1, but its function has been challenged recently. One other integral outer-membrane protein, GDAP1,
has also been implicated in Drp1-dependent mitochondrial division in mammals; however, the exact function of this protein awaits elucidation.
Several types of posttranslational modifications regulate Drp1 in mammals. Similar to mitofusins, the activity of Drp1 appears to be regulated by MITOL/March5-dependent
ubiquitination. In addition, SUMOylation, phosphorylation, and N-nitrosylation of Drp1 also control its functions.
Although Drp1 appears to bind directly to outer-membrane receptors to promote division in mammalian cells, Dnm1 is tethered to the outer membrane by two functionally
redundant protein complexes in yeast. Fis1 recruits Dnm1 via two WD40 domain-containing adaptor proteins, Mdv1 and Caf4. Parallel to this mechanism, Mdm36 and the cortical protein Num1 retain Dnm1 at the mitochondrial surface. The Num1-Mdm36 mechanism connects mitochondria to the cell cortex and promotes appropriate segregation and
inheritance of mitochondria during cytokinesis.
In algae, Dnm1 is proposed to associate with Fis1 and the WD40 domain-containing protein Mda1 on the outer membrane. Additionally, two proteins related to bacterial
division components, FtsZ and ZED, are located on the matrix side of the inner membrane and form a ring structure, which potentially mediates inner-membrane division. The
machinery necessary for inner-membrane division has yet to be identified in mammals and yeast.
Abbreviations
Mfn, mitofusin; G, GTPase; R, ring-domain; Ub, ubiquitin; P, phosphate; SUMO, small ubiquitin-like modifier; OM, outer membrane; IMS, intermembrane space; IM, inner membrane; SCF, Skp1-cullin-F box; MEF, mouse embryonic fibroblast. When the names for mammalian and their yeast orthologs differ, the yeast name is in parentheses.
Acknowledgments
We thank M. Iijima for helpful discussions and Y. Kageyama for providing the image of mitochondria in mouse embryonic fibroblasts. Y.T. is supported by a Japan Society for the
Promotion of Science fellowship. H.S. is supported by a National Institutes of Health grant (GM89853).
References
Campello, S., and Scorrano, L. (2010). Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 11, 678684.
Chang, C.R., and Blackstone, C. (2010). Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N Y Acad. Sci. 1201, 3439.
Chen, H., and Chan, D.C. (2009). Mitochondrial dynamicsfusion, fission, movement, and mitophagyin neurodegenerative diseases. Hum. Mol. Genet. 18 (R2), R169R176.
Hoppins, S., and Nunnari, J. (2009). The molecular mechanism of mitochondrial fusion. Biochim. Biophys. Acta 1793, 2026.
Kageyama, Y., Zhang, Z., and Sesaki, H. (2011). Mitochondrial division: molecular machinery and physiological functions. Curr. Opin. Cell Biol.
Kuroiwa, T. (2010). Mechanisms of organelle division and inheritance and their implications regarding the origin of eukaryotic cells. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 86,
455471.
McBride, H., and Soubannier, V. (2010). Mitochondrial function: OMA1 and OPA1, the grandmasters of mitochondrial health. Curr. Biol. 20, R274R276.
Palmer, C.S., Osellame, L.D., Laine, D., Koutsopoulos, O.S., Frazier, A.E., and Ryan, M.T. (2011). MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO
Rep. 12, 565573.
Westermann, B. (2010). Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin. Cell Dev. Biol.
21, 542549.
Youle, R.J., and Narendra, D.P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 914.
1158.e1 Cell 145, June 24, 2011 2011 Elsevier Inc. DOI 10.1016/j.cell.2011.06.018
You might also like
- Ebook American Corrections 13Th Edition Todd R Clear Online PDF All ChapterDocument69 pagesEbook American Corrections 13Th Edition Todd R Clear Online PDF All Chaptersonya.martinez866100% (11)
- Molecular Cell BiologyDocument94 pagesMolecular Cell BiologyPoorinku Poopinki94% (17)
- BTVS 5x16 "The Body" Audio CommentaryDocument12 pagesBTVS 5x16 "The Body" Audio CommentarynjzNo ratings yet
- Speech Acts: Are The Speaker's Utterances Which Convey Meaning and Make Listeners Do Specific Things (Austin, 1962)Document10 pagesSpeech Acts: Are The Speaker's Utterances Which Convey Meaning and Make Listeners Do Specific Things (Austin, 1962)Ashi Belmonte100% (4)
- Review Article: Mitochondrial Dynamics: Functional Link With ApoptosisDocument11 pagesReview Article: Mitochondrial Dynamics: Functional Link With ApoptosisAgnes GintingNo ratings yet
- Dysfunctional Mitochondrial Dynamics in The Pathophysiology of Neurodegenerative DiseasesDocument9 pagesDysfunctional Mitochondrial Dynamics in The Pathophysiology of Neurodegenerative DiseasesNuacha ZterezabiestNo ratings yet
- Mitochondria Fission and FusionDocument12 pagesMitochondria Fission and FusionAninditaDebPalNo ratings yet
- Pertumbuhan SelDocument37 pagesPertumbuhan SelbeniNo ratings yet
- 10 1242@jcs 203216Document50 pages10 1242@jcs 203216AnissaNo ratings yet
- TMP ACE6Document16 pagesTMP ACE6FrontiersNo ratings yet
- Mitophagy Parkinson PDFDocument7 pagesMitophagy Parkinson PDFAnonymous Sgxxu6No ratings yet
- NIH Public Access: Author ManuscriptDocument20 pagesNIH Public Access: Author ManuscriptTri SantosoNo ratings yet
- Mithocondrial Biogenesis by Exercise and Nutrients 2017 1357718365Document9 pagesMithocondrial Biogenesis by Exercise and Nutrients 2017 1357718365JE SnowNo ratings yet
- AMBRA1 Is Able To Induce Mitophagy Via LC3 Binding, Regardless of PARKIN and p62/SQSTM1Document14 pagesAMBRA1 Is Able To Induce Mitophagy Via LC3 Binding, Regardless of PARKIN and p62/SQSTM1Nikhil DevNo ratings yet
- PIIS009286741631652XDocument26 pagesPIIS009286741631652Xsumangal12995No ratings yet
- Renal Fibrosis: Not Just PAI-1 in The Sky: Agnes B. FogoDocument3 pagesRenal Fibrosis: Not Just PAI-1 in The Sky: Agnes B. FogoboxboxboxboxboxNo ratings yet
- Role and Mechanisms of Mitophagy in Liver DiseasesDocument30 pagesRole and Mechanisms of Mitophagy in Liver DiseasescarolinaNo ratings yet
- Phosphatidylinositol 3 - Kinase Signalling Supports Cell Height in Established Epithelial MonolayersDocument11 pagesPhosphatidylinositol 3 - Kinase Signalling Supports Cell Height in Established Epithelial MonolayersMohammed Elias AlamNo ratings yet
- Srep32933 PDFDocument13 pagesSrep32933 PDFCarolina RicárdezNo ratings yet
- Fcaa 135Document27 pagesFcaa 135Joao GomesNo ratings yet
- Fan2016 Tissue Repair and Regeneration PDFDocument15 pagesFan2016 Tissue Repair and Regeneration PDFDavid LuNo ratings yet
- Apoptosis (By Ayushi Maurya)Document20 pagesApoptosis (By Ayushi Maurya)Ayushi MauryaNo ratings yet
- Mitochondrial Dynamics in Type 2 Diabetes: Pathophysiological ImplicationsDocument13 pagesMitochondrial Dynamics in Type 2 Diabetes: Pathophysiological ImplicationsZaira Camila CriolloNo ratings yet
- Methamphetamine-Induced Occludin Endocytosis Is Mediated by Arp2-3 Complex Regulated Actin RearrangementDocument12 pagesMethamphetamine-Induced Occludin Endocytosis Is Mediated by Arp2-3 Complex Regulated Actin RearrangementKarina B Hernandez ANo ratings yet
- Bergo 2014Document16 pagesBergo 2014SARAHI MACIAS REALNo ratings yet
- Mitochondria More Than JustDocument10 pagesMitochondria More Than JustCarla MONo ratings yet
- tmp490 TMPDocument11 pagestmp490 TMPFrontiersNo ratings yet
- Full PDFDocument6 pagesFull PDFLateecka R KulkarniNo ratings yet
- Sakaeda2002 Article MDR1Up-RegulatedByApoptoticStiDocument7 pagesSakaeda2002 Article MDR1Up-RegulatedByApoptoticStiMarco BrithoNo ratings yet
- Molecular Mechanisms Underlying The Involvement of The S1R in Methamphetamine Mediated Microglia Polarization - Chao2017Document13 pagesMolecular Mechanisms Underlying The Involvement of The S1R in Methamphetamine Mediated Microglia Polarization - Chao2017nienminhNo ratings yet
- Assessing Mitochondria BiogenesisDocument7 pagesAssessing Mitochondria BiogenesisashueinNo ratings yet
- TMP 60 B5Document26 pagesTMP 60 B5FrontiersNo ratings yet
- Nutrient Regulation of MTORC1 at A Glance JCS 2019Document6 pagesNutrient Regulation of MTORC1 at A Glance JCS 2019Osvaldo VillarNo ratings yet
- 2017-Journal of NeurochemistryDocument87 pages2017-Journal of NeurochemistryFabio CampeottoNo ratings yet
- tmpC848 TMPDocument9 pagestmpC848 TMPFrontiersNo ratings yet
- ArticleDocument14 pagesArticleOusmane KoneNo ratings yet
- 10 1016@j Bbabio 2018 01 005Document7 pages10 1016@j Bbabio 2018 01 005Michi TristeNo ratings yet
- Mitochondrial Dysfunction and Mitophagy in Parkinson's: From Familial To Sporadic DiseaseDocument11 pagesMitochondrial Dysfunction and Mitophagy in Parkinson's: From Familial To Sporadic DiseaseKaycsa AdrianaNo ratings yet
- Artikel Prof Noriyuki KoibuchiDocument12 pagesArtikel Prof Noriyuki KoibuchiAndreas AdiwinataNo ratings yet
- Mclelland Et Al 2014 Parkin and Pink1 Function in A Vesicular Trafficking Pathway Regulating Mitochondrial QualityDocument14 pagesMclelland Et Al 2014 Parkin and Pink1 Function in A Vesicular Trafficking Pathway Regulating Mitochondrial QualityelenakocevaNo ratings yet
- Renal 3Document12 pagesRenal 3omdaNo ratings yet
- Chaperonas Moleculares PDFDocument11 pagesChaperonas Moleculares PDFMARIBELNo ratings yet
- tmpF47D TMPDocument15 pagestmpF47D TMPFrontiersNo ratings yet
- Stress Granule Formation Eif2a No Aumenta AtfDocument4 pagesStress Granule Formation Eif2a No Aumenta Atfb12loaljNo ratings yet
- 1 s2.0 S2352289524000535 MainDocument11 pages1 s2.0 S2352289524000535 MainMoo GeeNo ratings yet
- SIRT1 Promotes Cell Survival Under Stress by Deacetylation-Dependent Deactivation of Poly (ADP-Ribose) Polymerase 1Document15 pagesSIRT1 Promotes Cell Survival Under Stress by Deacetylation-Dependent Deactivation of Poly (ADP-Ribose) Polymerase 1SENTHILKUMAR B RAJAMOHANNo ratings yet
- TMP 857 FDocument11 pagesTMP 857 FFrontiersNo ratings yet
- The Role of Mitochondria in Apoptosis - PMCDocument34 pagesThe Role of Mitochondria in Apoptosis - PMCAnumol LoranceNo ratings yet
- Possible Mechanisms of Disease Development in Tuberous SclerosisDocument7 pagesPossible Mechanisms of Disease Development in Tuberous Sclerosisplastioid4079No ratings yet
- Acb 43 97Document13 pagesAcb 43 97maryjoseNo ratings yet
- (論文閱讀) 心理壓力導致生理疾病-壓力導致胃病Document15 pages(論文閱讀) 心理壓力導致生理疾病-壓力導致胃病徐No ratings yet
- Role of Apoptosis-Related Factors in Follicular Atresia: ReviewDocument10 pagesRole of Apoptosis-Related Factors in Follicular Atresia: ReviewLiliz D'Luph RamaNo ratings yet
- Transcription Physiology'' of Pigment Formation in Melanocytes: Central Role of MITFDocument11 pagesTranscription Physiology'' of Pigment Formation in Melanocytes: Central Role of MITFAbdur Rachman Ba'abdullahNo ratings yet
- ALS MechanismsDocument3 pagesALS MechanismsLiad ElmalemNo ratings yet
- TMP E85 CDocument39 pagesTMP E85 CFrontiersNo ratings yet
- ROCK - A549 MechanismDocument14 pagesROCK - A549 MechanismFriska Amanda Nur LailiyahNo ratings yet
- P53 and MitochondriaDocument9 pagesP53 and MitochondriaMouna RohanaNo ratings yet
- Macrophage Plasticity and Polarization in Vivo VeritasDocument9 pagesMacrophage Plasticity and Polarization in Vivo VeritasCristian Gutiérrez VeraNo ratings yet
- Book 2011Document102 pagesBook 2011fdsjklsdfxsdfsdfNo ratings yet
- tmpC7CF TMPDocument12 pagestmpC7CF TMPFrontiersNo ratings yet
- Involvement of Actin and Actin-Binding Proteins in CarcinogenesisDocument20 pagesInvolvement of Actin and Actin-Binding Proteins in CarcinogenesisVeronica JanethNo ratings yet
- tmpE4F4 TMPDocument13 pagestmpE4F4 TMPFrontiersNo ratings yet
- The Significance of The Metabolism of The Neurohormone MelatoninDocument11 pagesThe Significance of The Metabolism of The Neurohormone MelatoninDaniel Díaz CarrascoNo ratings yet
- Malate Aspartate ShuttleDocument19 pagesMalate Aspartate ShuttleGiuliano Ciolacu100% (1)
- IPC - Serii de DateDocument1 pageIPC - Serii de DateGiuliano CiolacuNo ratings yet
- Pi Is 0092867411008932Document3 pagesPi Is 0092867411008932Giuliano CiolacuNo ratings yet
- Janine Zieg, Paul L. Greer, and Michael E. Greenberg Department of Neurobiology, Harvard Medical School, Boston, MA 02115, USADocument3 pagesJanine Zieg, Paul L. Greer, and Michael E. Greenberg Department of Neurobiology, Harvard Medical School, Boston, MA 02115, USAGiuliano CiolacuNo ratings yet
- A 165185Document137 pagesA 165185Giuliano Ciolacu100% (1)
- On The Universality of The Riemann Zeta-FunctionDocument70 pagesOn The Universality of The Riemann Zeta-FunctionGiuliano Ciolacu100% (1)
- Neuron-Specific Protein Interactions of Drosophila CASK-are Revealed by Mass SpectrometryDocument10 pagesNeuron-Specific Protein Interactions of Drosophila CASK-are Revealed by Mass SpectrometryGiuliano CiolacuNo ratings yet
- Great Leaders Who Make The Mix WorkDocument7 pagesGreat Leaders Who Make The Mix WorkGiuliano CiolacuNo ratings yet
- Profilul Economic Al LuxemburguluiDocument103 pagesProfilul Economic Al LuxemburguluiGiuliano CiolacuNo ratings yet
- Lewis StructureDocument5 pagesLewis StructureGiuliano CiolacuNo ratings yet
- Chemistry - GlossaryDocument18 pagesChemistry - GlossaryGiuliano CiolacuNo ratings yet
- PHILOSOPHY 11 Q1 Week5 - 6 MELC7 8 MOD Mabuti Elizabeth and Obrero Daniel Daniel ObreroDocument19 pagesPHILOSOPHY 11 Q1 Week5 - 6 MELC7 8 MOD Mabuti Elizabeth and Obrero Daniel Daniel ObreroWilma DamoNo ratings yet
- AR-Raheeq Al-Makhtum (The Seald Nectar)Document297 pagesAR-Raheeq Al-Makhtum (The Seald Nectar)rabtay100% (1)
- Biography of Helen Keller - TNPSC General English by WWW - Tnpsc.academyDocument22 pagesBiography of Helen Keller - TNPSC General English by WWW - Tnpsc.academyMeenachisundaram.j BabuNo ratings yet
- GnosticismDocument3 pagesGnosticismGinaPraysNo ratings yet
- Tanween (Nunation) of The Arabic Nouns in 25 Sayings of Prophet Muhammad (PBUH)Document22 pagesTanween (Nunation) of The Arabic Nouns in 25 Sayings of Prophet Muhammad (PBUH)Jonathan MorganNo ratings yet
- Exposed Brickwork ConstructionDocument12 pagesExposed Brickwork ConstructionUzair Ahmed0% (1)
- Recepccion Documentos Aprendicces Logistica Empresarial 2018Document6 pagesRecepccion Documentos Aprendicces Logistica Empresarial 2018nasly castro garciaNo ratings yet
- What Are The Rules Regarding The Acceptance of A Proposal? Describe Them in Details.Document2 pagesWhat Are The Rules Regarding The Acceptance of A Proposal? Describe Them in Details.sunnykapoor7No ratings yet
- Nojpetén: EtymologyDocument6 pagesNojpetén: EtymologyOrgito LekaNo ratings yet
- Pakistan Stock Exchange Limited: List of Trading Right Entitlement (Tre) Certificate HoldersDocument36 pagesPakistan Stock Exchange Limited: List of Trading Right Entitlement (Tre) Certificate HoldersMuhammad AbidNo ratings yet
- Carlos Runcie-Tanaka: The Passion and Pulse of ClayDocument4 pagesCarlos Runcie-Tanaka: The Passion and Pulse of ClayScolli Huaranga GalarzaNo ratings yet
- 7 Doh Approved Herbal Medicine: Pictures Indicataion Contraindication Nursing ResponsibilityDocument5 pages7 Doh Approved Herbal Medicine: Pictures Indicataion Contraindication Nursing ResponsibilityShaira Ann CalambaNo ratings yet
- The Most Amazing Structure On EarthDocument2 pagesThe Most Amazing Structure On EarthStrafalogea SerbanNo ratings yet
- Narrative Medicine Form Function and EthicsDocument5 pagesNarrative Medicine Form Function and EthicsPetruta FlangeaNo ratings yet
- Cornell NotesDocument2 pagesCornell Notesgeophrey kajokiNo ratings yet
- Sanidad vs. COMELECDocument1 pageSanidad vs. COMELECDonna de RomaNo ratings yet
- Accessible Stations Subway MapDocument1 pageAccessible Stations Subway Mapmary Carmen FigueroaNo ratings yet
- 2021 Florida Complex League Schedule - REVISED 6.3.21Document4 pages2021 Florida Complex League Schedule - REVISED 6.3.21Josh Norris100% (1)
- EOU UNIT 11 - Đáp Án Eou 11Document8 pagesEOU UNIT 11 - Đáp Án Eou 11Trường Sơn NguyễnNo ratings yet
- Shelf-Life Expiration Date (SLED) in SAP MM.Document26 pagesShelf-Life Expiration Date (SLED) in SAP MM.maniNo ratings yet
- # 112 Change Solenoid On Top Drive1Document2 pages# 112 Change Solenoid On Top Drive1HSE ManagerNo ratings yet
- MIgrant Workers To PrisonersDocument57 pagesMIgrant Workers To PrisonersPrincess Janine SyNo ratings yet
- 2023 Tiger Neo Product Introduction-2023-4-25Document28 pages2023 Tiger Neo Product Introduction-2023-4-25xNooMx NajaNo ratings yet
- 1A Grammar Question Formation PDFDocument4 pages1A Grammar Question Formation PDFTijana CurcicNo ratings yet
- Bly, Robert (Ed.) - Friends, You Drank Some Darkness (Beacon, 1975)Document281 pagesBly, Robert (Ed.) - Friends, You Drank Some Darkness (Beacon, 1975)Mats CederNo ratings yet
- 3844 D DANQUAH Conditional Offer Letter 1Document3 pages3844 D DANQUAH Conditional Offer Letter 1Lozo DreNo ratings yet
- Group30 Assignment 1Document7 pagesGroup30 Assignment 1Rajat GargNo ratings yet