Professional Documents
Culture Documents
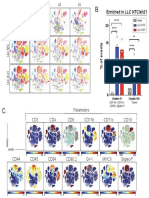
Supplementary Fig. 1: CD4+CD25+
Supplementary Fig. 1: CD4+CD25+
Uploaded by
Arnab DasCopyright:
Available Formats
You might also like
- Bearing Capacity Excel SheetDocument2 pagesBearing Capacity Excel SheetHanafiahHamzahNo ratings yet
- DESADV MapDocument3 pagesDESADV Maprubenmun_100% (3)
- SUE JOhnsonDocument72 pagesSUE JOhnsonannarudolf78100% (5)
- Cialis Case PDFDocument4 pagesCialis Case PDFinterpon07No ratings yet
- Sharon Packer - Movies and Modern PsycheDocument217 pagesSharon Packer - Movies and Modern PsycheZoran Kosjerina100% (2)
- Bumanlag, Paul Brian J. Cbet 19-503P Percentiles, Quartiles and Deciles For Grouped DataDocument2 pagesBumanlag, Paul Brian J. Cbet 19-503P Percentiles, Quartiles and Deciles For Grouped DataPaul Brian BumanlagNo ratings yet
- 13 Apr 2024Document1 page13 Apr 2024sohba2000No ratings yet
- LIA 001588 19.gcdDocument1 pageLIA 001588 19.gcdAnonymous GdWMlV46bUNo ratings yet
- Lab. Kimia Organik - Fmipa - UgmDocument1 pageLab. Kimia Organik - Fmipa - UgmPandji ZFNo ratings yet
- RND Systems Bcells BRDocument16 pagesRND Systems Bcells BRchernishovadoNo ratings yet
- Rpi Cheat Sheet: Main Program (.Text) : U V W X y Z (Document10 pagesRpi Cheat Sheet: Main Program (.Text) : U V W X y Z (Pablo Perez MartinNo ratings yet
- Cyclone CalculatorDocument20 pagesCyclone Calculatorhardik033No ratings yet
- Craig Chronic Lymphoid Disorders Useful Information CCEN India 2018 FinalDocument9 pagesCraig Chronic Lymphoid Disorders Useful Information CCEN India 2018 FinalBrahmananda ChakrabortyNo ratings yet
- Ethanol Sampel BDocument1 pageEthanol Sampel BAndika Rifqi RayendraNo ratings yet
- Hexano GCDDocument1 pageHexano GCDAnonymous GdWMlV46bUNo ratings yet
- Granger Causality-EAIIDocument10 pagesGranger Causality-EAIISNo ratings yet
- Oswal Hasil StatistikDocument17 pagesOswal Hasil StatistikBitpro disnakNTTNo ratings yet
- PID Pole Placement ControllerDocument16 pagesPID Pole Placement ControllerFiras BackourNo ratings yet
- Session XX-classDocument9 pagesSession XX-classswaroop shettyNo ratings yet
- Time, Clocks, and The Ordering of Events in A Distributed SystemDocument20 pagesTime, Clocks, and The Ordering of Events in A Distributed SystemAzeem TahirNo ratings yet
- CD1700/1800 References:: New FactorDocument5 pagesCD1700/1800 References:: New FactorfreedNo ratings yet
- Hematolog Controls LOT 0160Document3 pagesHematolog Controls LOT 0160jbNo ratings yet
- Activity 1 Finals CastorDocument4 pagesActivity 1 Finals CastorChona CastorNo ratings yet
- Ethanol Sampel ADocument1 pageEthanol Sampel AAndika Rifqi RayendraNo ratings yet
- SE170291Document2 pagesSE170291Nguyen Trong Tan (K17 HCM)No ratings yet
- Laboratory Records VARIANT II Beta Thal Short Program Le Lamentin 1 54Document3 pagesLaboratory Records VARIANT II Beta Thal Short Program Le Lamentin 1 54bassam alharaziNo ratings yet
- M1 14-AZ2 PENROSE Part 1 (Analysis) - BlankDocument5 pagesM1 14-AZ2 PENROSE Part 1 (Analysis) - BlankKhushi singhalNo ratings yet
- Chapter 16: Tool Kit For Working Capital ManagementDocument23 pagesChapter 16: Tool Kit For Working Capital ManagementosamaNo ratings yet
- Image Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceDocument5 pagesImage Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceFabian de Jesus Orozco MartinezNo ratings yet
- Image Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceDocument5 pagesImage Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceFabian de Jesus Orozco MartinezNo ratings yet
- Bullock Et Al 2018, Supplemental Figure 5Document1 pageBullock Et Al 2018, Supplemental Figure 5Abby KimballNo ratings yet
- Logam Berat CD Detergen: Logam Berat Cadmium (CD) Kons J.ikan J.mat %mortalitas Log K Probit LC50 (X) LC50Document4 pagesLogam Berat CD Detergen: Logam Berat Cadmium (CD) Kons J.ikan J.mat %mortalitas Log K Probit LC50 (X) LC50DianHardiantiNo ratings yet
- Table 2 Liver Cyt OxydaseDocument2 pagesTable 2 Liver Cyt OxydaseElias LawalNo ratings yet
- Activity No. 8 July 9, 2021 10:00 A.M. - 1:00 P.M. Submit This Activity Based On The Time Frame GivenDocument3 pagesActivity No. 8 July 9, 2021 10:00 A.M. - 1:00 P.M. Submit This Activity Based On The Time Frame GivenRosbert Cabug TulodNo ratings yet
- Supporting Information: Wakamatsu Et Al. 10.1073/pnas.1220688110Document4 pagesSupporting Information: Wakamatsu Et Al. 10.1073/pnas.1220688110Roger FigueiredoNo ratings yet
- BT02Document10 pagesBT02Trang Phạm ThịNo ratings yet
- Dap An Dieu Khien Qua Trinh PCTR421929Document3 pagesDap An Dieu Khien Qua Trinh PCTR421929Phi Cao TanNo ratings yet
- Predicine - Cancer - Multi Gene cfDNA Liquid Biopsy PanelDocument6 pagesPredicine - Cancer - Multi Gene cfDNA Liquid Biopsy PanelsagarkarvandeNo ratings yet
- Clients Order 2021Document135 pagesClients Order 2021Samuel LoyolaNo ratings yet
- Esquema Do ECS I38II1Document37 pagesEsquema Do ECS I38II1Wando MenezesNo ratings yet
- Take Home ExamDocument6 pagesTake Home ExamomarkezzalarabNo ratings yet
- Politeknik Kesehatan BandungDocument1 pagePoliteknik Kesehatan BandungAfni NurbayaniNo ratings yet
- Table of Blood Group Systems v. 9.0 03-FEB-2021Document3 pagesTable of Blood Group Systems v. 9.0 03-FEB-2021AnthonyNo ratings yet
- No. System Name System Symbol Gene Name(s) Number of Antigens Chromosomal Location CD NumbersDocument3 pagesNo. System Name System Symbol Gene Name(s) Number of Antigens Chromosomal Location CD NumbersAnthonyNo ratings yet
- Task 3 - Model AnswerDocument4 pagesTask 3 - Model AnswerHarry SinghNo ratings yet
- EuroFlow Ab Panels and Technical Information - Version 1.10Document14 pagesEuroFlow Ab Panels and Technical Information - Version 1.10Jéssica ParenteNo ratings yet
- KH2020Document513 pagesKH2020Hebert BallesterosNo ratings yet
- 4Document17 pages4M SiddiqNo ratings yet
- Gofast - Arm - Keil - FloatingPoint - ExecutionTimeDocument2 pagesGofast - Arm - Keil - FloatingPoint - ExecutionTimeDriveNo ratings yet
- HilmahDocument1 pageHilmahRSCERIA KANDANGANNo ratings yet
- Composition of EuroFlow Panels and Technical Information On Reagents Version 1.4Document14 pagesComposition of EuroFlow Panels and Technical Information On Reagents Version 1.4Liliana SolariNo ratings yet
- C LabSolutions Data Project1 ETO H-8Document1 pageC LabSolutions Data Project1 ETO H-8khizer iqbalNo ratings yet
- KH2020 Nucleus PDFDocument510 pagesKH2020 Nucleus PDFmrservice7782100% (1)
- Gas Flow Rate Calculation: Write in Green Cells OnlyDocument19 pagesGas Flow Rate Calculation: Write in Green Cells OnlymolcitoNo ratings yet
- Trabajo 2 EconometriaDocument6 pagesTrabajo 2 EconometriaJose Hernández CumplidoNo ratings yet
- Gmo Plant DDCQ 100% Gmo Non-Gmo Produc Ta Produc TBDocument4 pagesGmo Plant DDCQ 100% Gmo Non-Gmo Produc Ta Produc TBClaudia PetreNo ratings yet
- Ami A02Document1 pageAmi A02Anggun Pulihana WNo ratings yet
- Ethanol STD 3 PRSNDocument1 pageEthanol STD 3 PRSNAndika Rifqi RayendraNo ratings yet
- GhatgePatilGraph PDFDocument1 pageGhatgePatilGraph PDFbhageshlNo ratings yet
- Petrophysical Report 1Document5 pagesPetrophysical Report 1nashat90No ratings yet
- Changing Mal Treatment Policy-Prof L SalakoDocument17 pagesChanging Mal Treatment Policy-Prof L Salakoapi-3705046No ratings yet
- Phase 2 Infill Results Continue To Return High-Grade Over Broad Intervals With 25.3m at 14g/tDocument5 pagesPhase 2 Infill Results Continue To Return High-Grade Over Broad Intervals With 25.3m at 14g/ttomNo ratings yet
- Principles and Applications of Thermal AnalysisFrom EverandPrinciples and Applications of Thermal AnalysisPaul GabbottRating: 4 out of 5 stars4/5 (1)
- Chapter 37Document11 pagesChapter 37Amir MontanoNo ratings yet
- Prevention Practice: Dr. Sana Farooq BSPT, MSPT, DPT.DDocument26 pagesPrevention Practice: Dr. Sana Farooq BSPT, MSPT, DPT.DAnnie KhanNo ratings yet
- Lucina TechSheetDocument2 pagesLucina TechSheetMiguel M. Melchor RodríguezNo ratings yet
- Clinical Leadership Doc by MckinseyAug08Document32 pagesClinical Leadership Doc by MckinseyAug08Tyka Asta AmaliaNo ratings yet
- DEATH IN THE FREEZER ActivitiesDocument5 pagesDEATH IN THE FREEZER ActivitiesLaura Alicia TerragnoNo ratings yet
- Antiphospholipid Antibody Syndrome and PregnancyDocument17 pagesAntiphospholipid Antibody Syndrome and Pregnancybidan22No ratings yet
- Schizophrenia Lecture NotesDocument9 pagesSchizophrenia Lecture NotesDale Buckman100% (1)
- Pt. Dhainako Putra Sejati: Daftar Harga Jual BarangDocument16 pagesPt. Dhainako Putra Sejati: Daftar Harga Jual BarangBambang TriyonoNo ratings yet
- The 25 PROTOCOL: Important Exercise For SciaticaDocument8 pagesThe 25 PROTOCOL: Important Exercise For SciaticaDr Ahmed NabilNo ratings yet
- The Tangled Story of Alois AlzheimerDocument3 pagesThe Tangled Story of Alois AlzheimerJorge Arturo Cantu TorresNo ratings yet
- Characteristics of NewbornDocument102 pagesCharacteristics of NewbornSusan HepziNo ratings yet
- Extraocular Muscles Anatomy PDFDocument2 pagesExtraocular Muscles Anatomy PDFCherylNo ratings yet
- n378.008 Iris Website Staging of CKD PDFDocument8 pagesn378.008 Iris Website Staging of CKD PDFrutebeufNo ratings yet
- Micro X LPSDocument9 pagesMicro X LPSMarco Vinicio Lopez DelgadoNo ratings yet
- Hiaro Ser-05 20190528-1Document5 pagesHiaro Ser-05 20190528-1Prasad PNo ratings yet
- IWGDF Guidelines On The Prevention and Management of Diabetic Foot Disease 2019Document47 pagesIWGDF Guidelines On The Prevention and Management of Diabetic Foot Disease 2019Sidan EmozieNo ratings yet
- Rheumatic Fever: Assoc - Prof.Dr - Zurkurnai Yusof USMDocument25 pagesRheumatic Fever: Assoc - Prof.Dr - Zurkurnai Yusof USMfadlicardio100% (1)
- MANAGEMENT OF VERTICAL MAXILLARY EXCESS WWWDocument24 pagesMANAGEMENT OF VERTICAL MAXILLARY EXCESS WWWsweetieNo ratings yet
- Utas StudentsDocument2 pagesUtas StudentsJeffrey XieNo ratings yet
- My Mothers StoryDocument6 pagesMy Mothers Storybde_gnasNo ratings yet
- RyanDocument2,486 pagesRyanlunafranca22No ratings yet
- Affiniti 70 Spezifikation EnglischDocument40 pagesAffiniti 70 Spezifikation Englischias2008No ratings yet
- Sensory Processing DisorderDocument6 pagesSensory Processing DisorderMichael MoyalNo ratings yet
- Groper's ApplianceDocument4 pagesGroper's Appliancejinny1_0No ratings yet
- Cmo 14Document125 pagesCmo 14pirmiadi100% (1)
- Curriculam Vitae: ObjectiveDocument5 pagesCurriculam Vitae: ObjectiveGokul RajNo ratings yet
- Yoga Asanas For Neck Pain ReliefDocument5 pagesYoga Asanas For Neck Pain ReliefsbarathiNo ratings yet
Supplementary Fig. 1: CD4+CD25+
Supplementary Fig. 1: CD4+CD25+
Uploaded by
Arnab DasOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Supplementary Fig. 1: CD4+CD25+
Supplementary Fig. 1: CD4+CD25+
Uploaded by
Arnab DasCopyright:
Available Formats
Supplementary Fig.
a CD4+CD25+
LN Spleen
21.3 27.4
Events
Blimp1-GFP
b c
25 16
Foxp3 expression (x 103)
20
12
Tgfb1 expression
15
8
10
4
5
0 0
CD25+ CD25- CD25+ CD25-
GFP- GFP+ CD62L- CD62L+ GFP- GFP+ CD62L- CD62L+
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 2
a gfp/gfp (CD4+CD25+)
b gfp/gfp (CD4+CD25+)
75.3 18.9 48.8 2.4 Spleen Lymph node
GFP- 95.1 96.4
Events
CD62L
CD44
5.8 0.02 31.5 18.4
33.1 7.9 70.6 17.6
GFP+ 96.1 92.8
Events
CD69
GITR
48.3 10.7 10.3 1.5
25.3 8.1 28.7 24.5 Foxp3
CD38
IL-7R
58.1 8.4 44.7 2.0
Blimp1-GFP
c Ly5.1 : Blimp1gfp/gfp (CD4+) CD4+Foxp3+
+/+ (Ly5.1+) gfp/gfp (Ly5.2+) +/+ (Ly5.1+) gfp/gfp (Ly5.2+)
11.1 5.7 9.2 4.3 24.2 8.9 15.8 19.7
CD103
CD25
70.6 12.6 73.3 13.1 57.4 9.5 43.6 21.0
CTLA4 ICOS
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 3
a b 4.0
3.0
3.5
*
2.5
gfp/gfp Treg contribution (Log2)
Il10 expression (x103)
3.0
2.0 2.5
2.0
1.5 * *
1.5
1.0
1.0 *
0.5
0.5 n.s.
0 0.0
p
p
gf
gf
gf
gf
p/
p/
p/
p/
-0.5
+
+
+/
+/
+/
+/
gf
gf
gf
gf
LN spleen PP IEL Lung
CD25+ CD25+ CD25- CD25- -1.0
GFP+ GFP- CD62L- CD62L+
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 4
a
CD4+
PBS IL-2c
20.0 39.8
Events
Foxp3
b c IL-2 IL-2 + IL-4
IL-2 IL-2 + IL-4
78.9 89.7
Events
Events
IL-2 + IL-6 IL-2 + IL-12 IL-2 + IL-6 IL-2 + IL-12
79.1 79.1
Events
Events
Blimp1-GFP Foxp3
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 5
a b
CD4+ CD4+
mes LN Spleen mLN lung
13.8 7.1
Tbx21+/+ 5.31 1.15 4.94 0.73
Tbx21+/+
Blimp1+/gfp
91.5 1.90 90.8 3.51
Tbx21-/- 5.26 0.92 6.56 1.24 15.4 6.7
Tbx21-/-
Blimp1+/gfp
CD25
Foxp3
92.0 1.81 89.6 2.55
Blimp1-GFP CD4
c CD4+ Foxp3+ CD4+ Foxp3+
Tbx21+/+ Tbx21-/-
mLN lung mLN lung
18.8 7.51 26.5 9.15 19.6 8.8 21.7 5.36
CD103
22.7 50.9 29.0 35.3 28.6 43.0 43.1 29.8
CD62L CD62L
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 6
a spleen (CD4+)
+/+ Irf4-/-
14.4 0.7 6.8 0.01
Foxp3
81.6 0.3 93.2 0.01
KLRG1
b +/+ Irf4-/- Blimp1fl/flLck-Cre
(Ly5.1+) (Ly5.2+) (Ly5.2+)
39.7 0.3 0.7 0 19.5 0
T-bet
47.5 12.6 53.1 46.2 38.2 42.3
30 18.9 50.9 17.9 22.8 37.8
CD62L
49.9 1.2 30.6 0.5 37.8 1.7
CFSE
c CD4+
Tbx21+/+ Tbx21+/- Tbx21-/-
15.5 0.66 16.0 0.27 15.3 0.31
Foxp3
75.1 8.71 76.9 5.85 82.8 1.56
T-bet
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 7
a b Ly5.1 : Blimp1gfp/gfp
Ly5.1 : Blimp1+/+
CD4+Foxp3+ CD4+Foxp3+
20.6 20.5 7.8 4.2
+/+ mLN 70 +/+
Ly5.1 gfp/gfp
60
Ki67+ (% of Foxp3+)
50
9.8 49.1 51.5 36.6 40
25.8 35.4 9.9 8.6
+/+ 30
Lung
Ly5.2
20
CD103
10
Ki67
7.0 31.8 73.8 7.8
spleen LN lung
Bcl2 Ly5.1
c d Ly5.2+
CD4+ CD4+CD25+
43.7 21.1 40
Bim+/+ 14.5 2.1 Bim+/+ P=0.0006
Blimp1gfp/+ Blimp1gfp/+ 35
Blimp1-GFP+ (% of Tregs)
30
25
83.0 0.5 34.0 1.1
20
Bim-/- 22 6.1 Bim-/- 38.8 37.6
15
Blimp1gfp/+ Blimp1gfp/+
10
5
CD25
ICOS
70.3 1.7 21.2 2.4
+
-/-
+/
m
m
Blimp1-GFP Blimp1-GFP
Bi
Bi
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 8
a b
Ccr6 Klrg1
Il10
Blimp1
IRF4fl/+ / IRF4fl/-
c
IRF4fl/+ / IRF4fl/-
Blimp1+/gfp / Blimp1gfp/gfp
Blimp1+/gfp / Blimp1gfp/gfp
d
Blimp1 Ebi3 Ccr6 1.5
Rorc
2.0 2.0 4
Expression (x 102)
1.5 1.5 3
1.0
1.0 1.0 2
0.5
0.5 1.5 1
0 0 0 0
4+
4+
4+
4+
eg
eg
g
eg
g
g
g
D
D
e
D
D
re
re
re
re
Tr
Tr
Tr
Tr
C
C
C
C
-T
-T
-T
-T
+
ve
+
+
ve
ve
ve
FP
FP
FP
FP
FP
FP
FP
FP
ai
ai
ai
ai
G
G
G
N
N
N
200 Tcf7 80 Id3 10 Bcl2
8
e CD4+CD25+
Expression (x 102)
150 60
0.4 2.5
6
100 40
4
KLRG1
50 20
2
86.6 10.5
0 0 0
4+
4+
4+
eg
eg
eg
g
Blimp1-GFP
D
D
re
re
re
Tr
Tr
Tr
C
C
-T
-T
-T
+
+
e
ve
ve
FP
FP
FP
FP
FP
FP
v
ai
ai
ai
G
G
G
G
N
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 9
a CD4+
12.1 5.1
Foxp3
81.9 0.9
CD103
b +/+ Irf4-/- Irf4-/- Irf4-/-
empty vector empty vector Blimp1 vector IRF4 vector w/o
0.8 0.4 0.25 0.5 0.01
Foxp3
CD4
29.8 45.5 68 1.9 53.4 1.6 26.7 21.9
CD103
18.3 6.4 30.1 0 43.7 1.2 42.0 9.4
KLRG1
69.6 10.7 22.8 75.0
Events
ICOS
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 10
0.08
Irf4-/-
Il10 occupancy by IRF4 (% of input)
+/+
Blimp1fl/fl Lck-Cre
0.06
0.04
0.02
0.00
Pr S9
Pr S9
Pr S9
C S3
C S3
C S3
om
om
om
N
N
N
N
C
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 11
a
Il10 locus, intron 1
putative putative
IRF4 sites Blimp1 sites
mouse
mouse
human
human
b
Il10 locus, intron 1
IRF4 IRF4
Blimp1 Blimp1 IRF4
Blimp1 Blimp1 Blimp1
mouse
human
90 100 110 120 130 140 150 160 170 180 190
IRF4
Blimp1
mouse
human
200 210 220 230 240 250 260 270 280 290 300
Nature Immunology doi:10.1038/ni.2006
Supplementary Fig. 12
Nature Immunology doi:10.1038/ni.2006
Supplementary Table 1
Microarray analysis of GFP+ Treg cells from Blimp1gfp/gfp
and Blimp1+/gfp mice. Data is the fold change (Blimp1gfp/gfp / Blimp1+/gfp).
FDR
Nature Immunology doi:10.1038/ni.2006
Supplementary Table 2
Analysis of Blimp1-differentially expressed genes for their dependence on IRF4.
Data shows fold changes of these genes in the IRF4fl/- / IRF4fl/+ microarray
analysis (based on Zheng et al., 2009)17.
FDR
Nature Immunology doi:10.1038/ni.2006
Supplementary Material and Methods
RT-PCR primer sequences used for mRNA expression analysis:
Hprt: 5’- GGGGGCTATAAGTTCTTTGC-3’
5’- TCCAACACTTCGAGAGGTCC-3’
Foxp3: 5’- GGCCCTTCTCCAGGACAGA-3’
5’- GCTGATCATGGCTGGGTTGT-3’
Tgfb1: 5’- CAACGCCATCTATGAGAAAACC-3’
5’- AAGCCCTGTATTCCGTCTCC-3’
Il10: 5’- GAAGACCCTCAGGATGCGG-3’
5’- CCTGCTCCACTGCCTTGCT-3’
Tbx21: 5’- AGGGGACACTCGTATCAACAGA-3’
5’- AGGGGGCTTCCAACAATG-3’
Tcf7: 5’-AGCTTTCTCCACTCTACGAACA-3’
5’-AATCCAGAGAGATCGGGGGTC-3’
Id3: 5’-CTCTTAGCCTCTTGGACGACAT-3’
5’-GATCGAAGCTCATCCATGCCCT-3’
Ccr6: 5’-CCTGGGCAACATTATGGTGGT-3’
5’-CAGAACGGTAGGGTGAGGACA-3’
Rorc: 5’-TTTGGAACTGGCTTTCCATC-3’
5’-AAGATCTGCAGCTTTTCCACA-3’
Bcl2: 5’-TTATAAGCTGTCACAGAGGGGCTAC-3’
5’-GAACTCAAAGAAGGCCACAATCCTC-3’
RT-PCR primer sequences used for ChIP analysis:
Il10 CNS3 5’-AGTTGAGCCATTAGGGTTGGATAGAGG-3’
5’-ACCCCAGGCAGAAGTCAGTCTAATATC-3’
Nature Immunology doi:10.1038/ni.2006
Il10 CNS9 5’-CTTGAGGAAAAGCCAGCATC-3’
5’-TTTGCGTGTTCACCTGTGTT-3’
Il10 Prom 5’-GGCACCAGAACTCTCCTCTG-3’
5’-TGGGTTGAACGTCCGATATT-3’
Il10 PromA 5’-GACCTGGGAGTGCGTGAATGG-3’
5’-AGCGCTAAAGAACTGGTCGGAATG-3’
Il10 PromB 5’-TGCAGTAAGTACAGGTCACAGTCA-3’
5’-AGTTTTCTACCAGCAGCAAGCACA-3’
Il10 intron1 5’- CCCTCTTCTCATTCGTTTTCCA-3’
5’-CCAGGAAAGAATTTGAGAAAAATCA-3’
Ccr6 5’-GAACTCAGGATGGCGTGACTGGT-3’
5’-TCAATAACCCAGAGCACCGAGAA-3’
Cd19 3’ 5’- CTCGCCTAGCCTTTCTTC-3’
5’- TTCGGATGTGCCTGGGTTTAG-3’
Blimp1 3' 5’-ACCGTTGAAAGACGGTGACT-3’
5’-AATGGCCCACATCATCTGTG-3’
Blimp1 CNS9 5’-AAGCTGCTAAGTGGGAGAGT-3’
5’-GCATAATTTAGCGTTTGGTG-3’
Blimp1 Prom 5’-GCATGAGAGGCAGGGCAACA-3’
5’-CCTGCTAACTGAATACATTCAG-3’
Nature Immunology doi:10.1038/ni.2006
You might also like
- Bearing Capacity Excel SheetDocument2 pagesBearing Capacity Excel SheetHanafiahHamzahNo ratings yet
- DESADV MapDocument3 pagesDESADV Maprubenmun_100% (3)
- SUE JOhnsonDocument72 pagesSUE JOhnsonannarudolf78100% (5)
- Cialis Case PDFDocument4 pagesCialis Case PDFinterpon07No ratings yet
- Sharon Packer - Movies and Modern PsycheDocument217 pagesSharon Packer - Movies and Modern PsycheZoran Kosjerina100% (2)
- Bumanlag, Paul Brian J. Cbet 19-503P Percentiles, Quartiles and Deciles For Grouped DataDocument2 pagesBumanlag, Paul Brian J. Cbet 19-503P Percentiles, Quartiles and Deciles For Grouped DataPaul Brian BumanlagNo ratings yet
- 13 Apr 2024Document1 page13 Apr 2024sohba2000No ratings yet
- LIA 001588 19.gcdDocument1 pageLIA 001588 19.gcdAnonymous GdWMlV46bUNo ratings yet
- Lab. Kimia Organik - Fmipa - UgmDocument1 pageLab. Kimia Organik - Fmipa - UgmPandji ZFNo ratings yet
- RND Systems Bcells BRDocument16 pagesRND Systems Bcells BRchernishovadoNo ratings yet
- Rpi Cheat Sheet: Main Program (.Text) : U V W X y Z (Document10 pagesRpi Cheat Sheet: Main Program (.Text) : U V W X y Z (Pablo Perez MartinNo ratings yet
- Cyclone CalculatorDocument20 pagesCyclone Calculatorhardik033No ratings yet
- Craig Chronic Lymphoid Disorders Useful Information CCEN India 2018 FinalDocument9 pagesCraig Chronic Lymphoid Disorders Useful Information CCEN India 2018 FinalBrahmananda ChakrabortyNo ratings yet
- Ethanol Sampel BDocument1 pageEthanol Sampel BAndika Rifqi RayendraNo ratings yet
- Hexano GCDDocument1 pageHexano GCDAnonymous GdWMlV46bUNo ratings yet
- Granger Causality-EAIIDocument10 pagesGranger Causality-EAIISNo ratings yet
- Oswal Hasil StatistikDocument17 pagesOswal Hasil StatistikBitpro disnakNTTNo ratings yet
- PID Pole Placement ControllerDocument16 pagesPID Pole Placement ControllerFiras BackourNo ratings yet
- Session XX-classDocument9 pagesSession XX-classswaroop shettyNo ratings yet
- Time, Clocks, and The Ordering of Events in A Distributed SystemDocument20 pagesTime, Clocks, and The Ordering of Events in A Distributed SystemAzeem TahirNo ratings yet
- CD1700/1800 References:: New FactorDocument5 pagesCD1700/1800 References:: New FactorfreedNo ratings yet
- Hematolog Controls LOT 0160Document3 pagesHematolog Controls LOT 0160jbNo ratings yet
- Activity 1 Finals CastorDocument4 pagesActivity 1 Finals CastorChona CastorNo ratings yet
- Ethanol Sampel ADocument1 pageEthanol Sampel AAndika Rifqi RayendraNo ratings yet
- SE170291Document2 pagesSE170291Nguyen Trong Tan (K17 HCM)No ratings yet
- Laboratory Records VARIANT II Beta Thal Short Program Le Lamentin 1 54Document3 pagesLaboratory Records VARIANT II Beta Thal Short Program Le Lamentin 1 54bassam alharaziNo ratings yet
- M1 14-AZ2 PENROSE Part 1 (Analysis) - BlankDocument5 pagesM1 14-AZ2 PENROSE Part 1 (Analysis) - BlankKhushi singhalNo ratings yet
- Chapter 16: Tool Kit For Working Capital ManagementDocument23 pagesChapter 16: Tool Kit For Working Capital ManagementosamaNo ratings yet
- Image Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceDocument5 pagesImage Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceFabian de Jesus Orozco MartinezNo ratings yet
- Image Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceDocument5 pagesImage Name: 5 Accelerating Voltage: 15.0 KV Magnification: 3000 Detector: NanotraceFabian de Jesus Orozco MartinezNo ratings yet
- Bullock Et Al 2018, Supplemental Figure 5Document1 pageBullock Et Al 2018, Supplemental Figure 5Abby KimballNo ratings yet
- Logam Berat CD Detergen: Logam Berat Cadmium (CD) Kons J.ikan J.mat %mortalitas Log K Probit LC50 (X) LC50Document4 pagesLogam Berat CD Detergen: Logam Berat Cadmium (CD) Kons J.ikan J.mat %mortalitas Log K Probit LC50 (X) LC50DianHardiantiNo ratings yet
- Table 2 Liver Cyt OxydaseDocument2 pagesTable 2 Liver Cyt OxydaseElias LawalNo ratings yet
- Activity No. 8 July 9, 2021 10:00 A.M. - 1:00 P.M. Submit This Activity Based On The Time Frame GivenDocument3 pagesActivity No. 8 July 9, 2021 10:00 A.M. - 1:00 P.M. Submit This Activity Based On The Time Frame GivenRosbert Cabug TulodNo ratings yet
- Supporting Information: Wakamatsu Et Al. 10.1073/pnas.1220688110Document4 pagesSupporting Information: Wakamatsu Et Al. 10.1073/pnas.1220688110Roger FigueiredoNo ratings yet
- BT02Document10 pagesBT02Trang Phạm ThịNo ratings yet
- Dap An Dieu Khien Qua Trinh PCTR421929Document3 pagesDap An Dieu Khien Qua Trinh PCTR421929Phi Cao TanNo ratings yet
- Predicine - Cancer - Multi Gene cfDNA Liquid Biopsy PanelDocument6 pagesPredicine - Cancer - Multi Gene cfDNA Liquid Biopsy PanelsagarkarvandeNo ratings yet
- Clients Order 2021Document135 pagesClients Order 2021Samuel LoyolaNo ratings yet
- Esquema Do ECS I38II1Document37 pagesEsquema Do ECS I38II1Wando MenezesNo ratings yet
- Take Home ExamDocument6 pagesTake Home ExamomarkezzalarabNo ratings yet
- Politeknik Kesehatan BandungDocument1 pagePoliteknik Kesehatan BandungAfni NurbayaniNo ratings yet
- Table of Blood Group Systems v. 9.0 03-FEB-2021Document3 pagesTable of Blood Group Systems v. 9.0 03-FEB-2021AnthonyNo ratings yet
- No. System Name System Symbol Gene Name(s) Number of Antigens Chromosomal Location CD NumbersDocument3 pagesNo. System Name System Symbol Gene Name(s) Number of Antigens Chromosomal Location CD NumbersAnthonyNo ratings yet
- Task 3 - Model AnswerDocument4 pagesTask 3 - Model AnswerHarry SinghNo ratings yet
- EuroFlow Ab Panels and Technical Information - Version 1.10Document14 pagesEuroFlow Ab Panels and Technical Information - Version 1.10Jéssica ParenteNo ratings yet
- KH2020Document513 pagesKH2020Hebert BallesterosNo ratings yet
- 4Document17 pages4M SiddiqNo ratings yet
- Gofast - Arm - Keil - FloatingPoint - ExecutionTimeDocument2 pagesGofast - Arm - Keil - FloatingPoint - ExecutionTimeDriveNo ratings yet
- HilmahDocument1 pageHilmahRSCERIA KANDANGANNo ratings yet
- Composition of EuroFlow Panels and Technical Information On Reagents Version 1.4Document14 pagesComposition of EuroFlow Panels and Technical Information On Reagents Version 1.4Liliana SolariNo ratings yet
- C LabSolutions Data Project1 ETO H-8Document1 pageC LabSolutions Data Project1 ETO H-8khizer iqbalNo ratings yet
- KH2020 Nucleus PDFDocument510 pagesKH2020 Nucleus PDFmrservice7782100% (1)
- Gas Flow Rate Calculation: Write in Green Cells OnlyDocument19 pagesGas Flow Rate Calculation: Write in Green Cells OnlymolcitoNo ratings yet
- Trabajo 2 EconometriaDocument6 pagesTrabajo 2 EconometriaJose Hernández CumplidoNo ratings yet
- Gmo Plant DDCQ 100% Gmo Non-Gmo Produc Ta Produc TBDocument4 pagesGmo Plant DDCQ 100% Gmo Non-Gmo Produc Ta Produc TBClaudia PetreNo ratings yet
- Ami A02Document1 pageAmi A02Anggun Pulihana WNo ratings yet
- Ethanol STD 3 PRSNDocument1 pageEthanol STD 3 PRSNAndika Rifqi RayendraNo ratings yet
- GhatgePatilGraph PDFDocument1 pageGhatgePatilGraph PDFbhageshlNo ratings yet
- Petrophysical Report 1Document5 pagesPetrophysical Report 1nashat90No ratings yet
- Changing Mal Treatment Policy-Prof L SalakoDocument17 pagesChanging Mal Treatment Policy-Prof L Salakoapi-3705046No ratings yet
- Phase 2 Infill Results Continue To Return High-Grade Over Broad Intervals With 25.3m at 14g/tDocument5 pagesPhase 2 Infill Results Continue To Return High-Grade Over Broad Intervals With 25.3m at 14g/ttomNo ratings yet
- Principles and Applications of Thermal AnalysisFrom EverandPrinciples and Applications of Thermal AnalysisPaul GabbottRating: 4 out of 5 stars4/5 (1)
- Chapter 37Document11 pagesChapter 37Amir MontanoNo ratings yet
- Prevention Practice: Dr. Sana Farooq BSPT, MSPT, DPT.DDocument26 pagesPrevention Practice: Dr. Sana Farooq BSPT, MSPT, DPT.DAnnie KhanNo ratings yet
- Lucina TechSheetDocument2 pagesLucina TechSheetMiguel M. Melchor RodríguezNo ratings yet
- Clinical Leadership Doc by MckinseyAug08Document32 pagesClinical Leadership Doc by MckinseyAug08Tyka Asta AmaliaNo ratings yet
- DEATH IN THE FREEZER ActivitiesDocument5 pagesDEATH IN THE FREEZER ActivitiesLaura Alicia TerragnoNo ratings yet
- Antiphospholipid Antibody Syndrome and PregnancyDocument17 pagesAntiphospholipid Antibody Syndrome and Pregnancybidan22No ratings yet
- Schizophrenia Lecture NotesDocument9 pagesSchizophrenia Lecture NotesDale Buckman100% (1)
- Pt. Dhainako Putra Sejati: Daftar Harga Jual BarangDocument16 pagesPt. Dhainako Putra Sejati: Daftar Harga Jual BarangBambang TriyonoNo ratings yet
- The 25 PROTOCOL: Important Exercise For SciaticaDocument8 pagesThe 25 PROTOCOL: Important Exercise For SciaticaDr Ahmed NabilNo ratings yet
- The Tangled Story of Alois AlzheimerDocument3 pagesThe Tangled Story of Alois AlzheimerJorge Arturo Cantu TorresNo ratings yet
- Characteristics of NewbornDocument102 pagesCharacteristics of NewbornSusan HepziNo ratings yet
- Extraocular Muscles Anatomy PDFDocument2 pagesExtraocular Muscles Anatomy PDFCherylNo ratings yet
- n378.008 Iris Website Staging of CKD PDFDocument8 pagesn378.008 Iris Website Staging of CKD PDFrutebeufNo ratings yet
- Micro X LPSDocument9 pagesMicro X LPSMarco Vinicio Lopez DelgadoNo ratings yet
- Hiaro Ser-05 20190528-1Document5 pagesHiaro Ser-05 20190528-1Prasad PNo ratings yet
- IWGDF Guidelines On The Prevention and Management of Diabetic Foot Disease 2019Document47 pagesIWGDF Guidelines On The Prevention and Management of Diabetic Foot Disease 2019Sidan EmozieNo ratings yet
- Rheumatic Fever: Assoc - Prof.Dr - Zurkurnai Yusof USMDocument25 pagesRheumatic Fever: Assoc - Prof.Dr - Zurkurnai Yusof USMfadlicardio100% (1)
- MANAGEMENT OF VERTICAL MAXILLARY EXCESS WWWDocument24 pagesMANAGEMENT OF VERTICAL MAXILLARY EXCESS WWWsweetieNo ratings yet
- Utas StudentsDocument2 pagesUtas StudentsJeffrey XieNo ratings yet
- My Mothers StoryDocument6 pagesMy Mothers Storybde_gnasNo ratings yet
- RyanDocument2,486 pagesRyanlunafranca22No ratings yet
- Affiniti 70 Spezifikation EnglischDocument40 pagesAffiniti 70 Spezifikation Englischias2008No ratings yet
- Sensory Processing DisorderDocument6 pagesSensory Processing DisorderMichael MoyalNo ratings yet
- Groper's ApplianceDocument4 pagesGroper's Appliancejinny1_0No ratings yet
- Cmo 14Document125 pagesCmo 14pirmiadi100% (1)
- Curriculam Vitae: ObjectiveDocument5 pagesCurriculam Vitae: ObjectiveGokul RajNo ratings yet
- Yoga Asanas For Neck Pain ReliefDocument5 pagesYoga Asanas For Neck Pain ReliefsbarathiNo ratings yet