Professional Documents
Culture Documents
Bayesian Networks Applied To Modeling Cellular Networks
Bayesian Networks Applied To Modeling Cellular Networks
Uploaded by
Gürkan GülerOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Bayesian Networks Applied To Modeling Cellular Networks
Bayesian Networks Applied To Modeling Cellular Networks
Uploaded by
Gürkan GülerCopyright:
Available Formats
Bayesian Networks Applied to Modeling Cellular Networks
BMI/CS 576 www.biostat.wisc.edu/bmi576/ Mark Craven craven@biostat.wisc.edu Fall 2011
Bayesian networks
! a BN is a Directed Acyclic Graph (DAG) in which ! the nodes denote random variables ! each node X has a conditional probability distribution (CPD) representing P(X | Parents(X) ) ! the intuitive meaning of an arc from X to Y is that X directly influences Y ! formally: each variable X is independent of its nondescendants given its parents
Probabilistic model of lac operon
! suppose we represent the system by the following discrete variables L (lactose) present, absent G (glucose) present, absent I (lacI) present, absent C (CAP) present, absent I-active (lacI unbound) present, absent C-active (CAP bound) present, absent Z (lacZ) high, low, absent ! suppose (realistically) the system is not completely deterministic ! the joint distribution of the variables could be specified by 26 ! 3 = 192 parameters
A Bayesian network for the lac system
L I G C
I-active
C-active
A Bayesian network for the lac system
P(L)
absent 0.9 present 0.1
We also have CPDs for I, G, C, C-active
I-active C-active
P ( I-active | L, I )
L absent absent present present I absent present absent present absent 0.9 0.1 0.9 0.9 present 0.1 0.9 0.1 0.1 I-active absent absent present present
Z P ( Z | I-active, C-active )
C-active absent present absent present absent 0.1 0.1 0.8 0.8 low 0.8 0.1 0.1 0.1 high 0.1 0.8 0.1 0.1
Bayesian networks
! a BN provides a factored representation of the joint probability distribution
P(L,I,Ia,G,Ca,Z) = P(L) " P(I) " P(Ia | L,I) " P(G) " P(C) " P(Ca | G,C) " P(Z | Ia,Ca)
I-active
C-active
! this representation of the joint distribution can be specified with 36 parameters (vs. 192 for the unfactored representation)
Representing CPDs for discrete variables
! CPDs can be represented using tables or trees ! consider the following case with Boolean variables A, B, C, D
P( D | A, B, C ) P( D | A, B, C )
A t t t t f f f f B t t f f t t f f C t f t f t f t f t 0.9 0.9 0.9 0.9 0.8 0.5 0.5 0.5 f 0.1 0.1 0.1 0.1 0.2 0.5 0.5 0.5 f t f f
A
t
B
t
Pr(D = t) = 0.9
Pr(D = t) = 0.5
Pr(D = t) = 0.5
Pr(D = t) = 0.8
Representing CPDs for continuous variables
! we can also model the distribution of continuous variables in Bayesian networks ! one approach: linear Gaussian models
Pr(X | u1 ,...,uk ) ~ N(a0 + # ai " ui , $ 2 )
i
! X normally distributed around a mean that depends linearly on values of its parents ui!
P(X | u1 )
u1
The parameter learning task
! Given: a set of training instances, the graph structure of a BN
L L present present absent G present present present I present present present C present present present I-active absent absent present C-active absent absent absent Z low absent high I-active
C-active
...
Z
! Do: infer the parameters of the CPDs ! this is straightforward when there arent missing values, hidden variables
The structure learning task
! Given: a set of training instances
L present present absent G present present present I present present present C present present present I-active absent absent present C-active absent absent absent Z low absent high
...
! Do: infer the graph structure (and perhaps the parameters of the CPDs too)
You might also like
- Chapter 9 Data MiningDocument147 pagesChapter 9 Data MiningYUTRYU8TYURTNo ratings yet
- BayesianNetworks ReducedDocument14 pagesBayesianNetworks Reducedastir1234No ratings yet
- CS 391L: Machine Learning: Bayesian Learning: Beyond Naïve BayesDocument29 pagesCS 391L: Machine Learning: Bayesian Learning: Beyond Naïve BayesFaizabadi Ahmed RimazNo ratings yet
- Lecture 15: Bayesian Networks III: CS221 / Autumn 2015 / LiangDocument70 pagesLecture 15: Bayesian Networks III: CS221 / Autumn 2015 / LiangBillybob EdwardsNo ratings yet
- Bayesian NeworksDocument32 pagesBayesian NeworksSAMARTH CHAUHANNo ratings yet
- Statistical Methods For Bioinformatics Lecture 5Document48 pagesStatistical Methods For Bioinformatics Lecture 5javabe7544No ratings yet
- Introduc) On To Probabilis) C Latent Seman) C Analysis: NYP Predic) Ve Analy) Cs Meetup June 10, 2010Document26 pagesIntroduc) On To Probabilis) C Latent Seman) C Analysis: NYP Predic) Ve Analy) Cs Meetup June 10, 2010김형진No ratings yet
- Graphical Models - InferenceDocument13 pagesGraphical Models - InferenceNhơn Phạm ThànhNo ratings yet
- Bayesian NetworksDocument35 pagesBayesian NetworksIrfan HussainNo ratings yet
- Principal Component Analysis (PCA) - : San José State University Math 253: Mathematical Methods For Data VisualizationDocument49 pagesPrincipal Component Analysis (PCA) - : San José State University Math 253: Mathematical Methods For Data VisualizationAndreea IsarNo ratings yet
- 0 BN ProbabilityDocument11 pages0 BN ProbabilitySobanNo ratings yet
- Bayesian Networks 2011-01-02Document106 pagesBayesian Networks 2011-01-02vctwmsmeNo ratings yet
- HW3Document2 pagesHW3SNo ratings yet
- Exercises 02Document3 pagesExercises 02roxaneNo ratings yet
- Good BayesianNetworksPrimerDocument23 pagesGood BayesianNetworksPrimerpradnya kingeNo ratings yet
- Naïve Bayes Classifier: Dr. Hussain DawoodDocument20 pagesNaïve Bayes Classifier: Dr. Hussain DawoodQasim AbidNo ratings yet
- Sparse Principal Component AnalysisDocument30 pagesSparse Principal Component AnalysisInSilicoBiology BiologyNo ratings yet
- Minsky y PapertDocument77 pagesMinsky y PapertHenrry CastilloNo ratings yet
- Machine Learning 10-601, 10-301: TodayDocument44 pagesMachine Learning 10-601, 10-301: TodaySasanka Sekhar SahuNo ratings yet
- Class19 ApproxinfDocument45 pagesClass19 ApproxinfKhadija M.No ratings yet
- Bayesian Modelling II: Saad JbabdiDocument41 pagesBayesian Modelling II: Saad JbabdiMauricio Garcia FeriaNo ratings yet
- Naive BayesDocument31 pagesNaive Bayesakrab.tech7No ratings yet
- Naïve Bayes Classifier: Dr. Hussain DawoodDocument20 pagesNaïve Bayes Classifier: Dr. Hussain DawoodQasim AbidNo ratings yet
- All of Graphical ModelsDocument135 pagesAll of Graphical ModelsjoseluismbNo ratings yet
- NeurIPS 2021 Graph Posterior Network Bayesian Predictive Uncertainty For Node Classification PaperDocument16 pagesNeurIPS 2021 Graph Posterior Network Bayesian Predictive Uncertainty For Node Classification Paper최윤혁No ratings yet
- Analysis of Binary Panel Data by Static and Dynamic Logit ModelsDocument45 pagesAnalysis of Binary Panel Data by Static and Dynamic Logit ModelsKiko LeónNo ratings yet
- Final 2002Document18 pagesFinal 2002Muhammad MurtazaNo ratings yet
- CS 215: Data Analysis and Interpretation: Sample QuestionsDocument10 pagesCS 215: Data Analysis and Interpretation: Sample QuestionsVinayaka GosulaNo ratings yet
- Bayes Nets 2015 PDFDocument19 pagesBayes Nets 2015 PDFSandeep GuptaNo ratings yet
- Aaai Grltutorial Part1 NoderepsDocument58 pagesAaai Grltutorial Part1 NoderepsJishnu JayarajNo ratings yet
- Exercise Sheet 1: Quantum Information - Summer Semester 2020Document2 pagesExercise Sheet 1: Quantum Information - Summer Semester 2020Kiran AdhikariNo ratings yet
- Bayesian Networks: Independencies and Inference: Scott Davies and Andrew MooreDocument21 pagesBayesian Networks: Independencies and Inference: Scott Davies and Andrew MooreMusanif EfendiNo ratings yet
- Week2-Fuzzy Logic and ReasoningDocument48 pagesWeek2-Fuzzy Logic and ReasoningMohamed mohamedNo ratings yet
- Machine Learning and Pattern Recognition Week 10 - Bayes - Logistic - RegressionDocument4 pagesMachine Learning and Pattern Recognition Week 10 - Bayes - Logistic - RegressionzeliawillscumbergNo ratings yet
- Vision 7CameraModelA S09Document28 pagesVision 7CameraModelA S09Kartik DuttaNo ratings yet
- Positional Analysis of Multi-Mode Fuzzy Networks: Jelena Ignjatovi C, Ivan Stankovi CDocument25 pagesPositional Analysis of Multi-Mode Fuzzy Networks: Jelena Ignjatovi C, Ivan Stankovi CMiroslav ĆirićNo ratings yet
- Annotated 4 Ch4 Linear Regression F2014Document11 pagesAnnotated 4 Ch4 Linear Regression F2014Bob HopeNo ratings yet
- Statistical Machine Learning Solutions For Exam 2019-03-15Document5 pagesStatistical Machine Learning Solutions For Exam 2019-03-15Yu Ching LeeNo ratings yet
- K - Nearest Neighbours Classifier / RegressorDocument35 pagesK - Nearest Neighbours Classifier / RegressorSmitNo ratings yet
- Analysis of Complex Sample Survey DataDocument24 pagesAnalysis of Complex Sample Survey DataCamilo Gutiérrez PatiñoNo ratings yet
- Introduction To Bayesian Methods With An ExampleDocument25 pagesIntroduction To Bayesian Methods With An ExampleLucas RobertoNo ratings yet
- CS 229, Public Course Problem Set #4: Unsupervised Learning and Re-Inforcement LearningDocument5 pagesCS 229, Public Course Problem Set #4: Unsupervised Learning and Re-Inforcement Learningsuhar adiNo ratings yet
- Properties of The OLS Estimator: Quantitative Methods 2Document57 pagesProperties of The OLS Estimator: Quantitative Methods 2rendangziNo ratings yet
- Statistics by CG50 UpdatedDocument30 pagesStatistics by CG50 Updatedsetokaiba12No ratings yet
- Lecture 3: Directed Graphical Models and Latent Variables Based On Slides byDocument48 pagesLecture 3: Directed Graphical Models and Latent Variables Based On Slides byRohit PandeyNo ratings yet
- Lecture 7 - Integrated Analysis With RDocument79 pagesLecture 7 - Integrated Analysis With RAnurag LaddhaNo ratings yet
- SimLinearReg OPPDocument11 pagesSimLinearReg OPPank93753No ratings yet
- Bayesian Belief NetworkDocument10 pagesBayesian Belief NetworkRohan RathiNo ratings yet
- Methods of Knowledge Engineering Bayes Networks, Probabilistic ModelingDocument34 pagesMethods of Knowledge Engineering Bayes Networks, Probabilistic ModelingPavel ZinevkaNo ratings yet
- An Overview of Edward: A Probabilistic Programming System: Dustin Tran Columbia UniversityDocument34 pagesAn Overview of Edward: A Probabilistic Programming System: Dustin Tran Columbia UniversityStanislav SeNo ratings yet
- Mlmultiplelinearregression 170919114353 PDFDocument8 pagesMlmultiplelinearregression 170919114353 PDFNguyen Huu Hanh LamNo ratings yet
- Data Augmentation For Support Vector Machines: Mallick Et Al. 2005 George and Mcculloch 1993 1997 Ishwaran and Rao 2005Document24 pagesData Augmentation For Support Vector Machines: Mallick Et Al. 2005 George and Mcculloch 1993 1997 Ishwaran and Rao 2005EZ112No ratings yet
- Chapter 9 Bayes FuzzyDocument70 pagesChapter 9 Bayes FuzzyLe Phu TranNo ratings yet
- Runge Phenomenon: Guided By: Dr. Geetanjali PradhanDocument29 pagesRunge Phenomenon: Guided By: Dr. Geetanjali PradhanSwarupa SarangiNo ratings yet
- Graphical Models: Worksheet 2 MT 2020: A: Warm UpDocument4 pagesGraphical Models: Worksheet 2 MT 2020: A: Warm Upscribdaccount314159No ratings yet
- Differential CalculusDocument85 pagesDifferential CalculusParminder singh parmarNo ratings yet
- 6.867 Section 3: Classification: 1 Intro 2 2 Representation 2 3 Probabilistic Models 2Document10 pages6.867 Section 3: Classification: 1 Intro 2 2 Representation 2 3 Probabilistic Models 2Suchan KhankluayNo ratings yet
- Shulman 2022 04 28Document42 pagesShulman 2022 04 28detraf01No ratings yet
- Sparse Principal Component AnalysisDocument23 pagesSparse Principal Component Analysis方鑫然No ratings yet
- Engineering Visualization: Question BankDocument10 pagesEngineering Visualization: Question Bankloda lassanNo ratings yet
- Physics XI STBB MOC 2023Document3 pagesPhysics XI STBB MOC 2023Habib U Zaman MemonNo ratings yet
- Fieldlines LectnotesDocument15 pagesFieldlines LectnotesZulhadi Iskandar RadziNo ratings yet
- Quarterly Report On Rasa Basa ImplementationDocument2 pagesQuarterly Report On Rasa Basa ImplementationJonathan Olegario100% (2)
- Redox-Uat s3 Ap-South-1 Amazonaws combirla2019May20CkbckEJlUyx3UHaaL8A1 PDFDocument22 pagesRedox-Uat s3 Ap-South-1 Amazonaws combirla2019May20CkbckEJlUyx3UHaaL8A1 PDFAryan JainNo ratings yet
- Kemper Profiler MIDI DocumentationDocument15 pagesKemper Profiler MIDI DocumentationAlexandre NovaisNo ratings yet
- External Pressure Rating Calculation For Pressure Vessels PPD 10104 and 10105 Per ASME Section VIII, Division 1Document2 pagesExternal Pressure Rating Calculation For Pressure Vessels PPD 10104 and 10105 Per ASME Section VIII, Division 1Pratik JainNo ratings yet
- Minka Dirichlet PDFDocument14 pagesMinka Dirichlet PDFFarhan KhawarNo ratings yet
- A025, A031, A035 - Cylinder Spring DesignDocument10 pagesA025, A031, A035 - Cylinder Spring DesignYash ZankhariyaNo ratings yet
- Pure MTC Seminar Questions 2022Document6 pagesPure MTC Seminar Questions 2022Majanga JohnnyNo ratings yet
- Experimental Analysis of Water Distribution Network and Its Hydraulic Simulation by EpanetDocument8 pagesExperimental Analysis of Water Distribution Network and Its Hydraulic Simulation by EpanetFisehaNo ratings yet
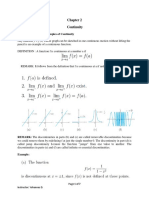
- Continuity: 2.1. Definition and Examples of ContinuityDocument7 pagesContinuity: 2.1. Definition and Examples of ContinuityBelew BelewNo ratings yet
- V4A Group2Document21 pagesV4A Group2Bane BarrionNo ratings yet
- Unit 2 Size ReductionDocument7 pagesUnit 2 Size ReductionKen GeronimoNo ratings yet
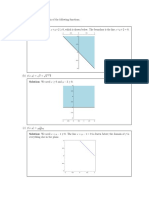
- Sketch The Domain of FunctionsDocument3 pagesSketch The Domain of FunctionsPedroNo ratings yet
- Simulation Methods in Statistical Physics, Project Report, FK7029, Stockholm UniversityDocument31 pagesSimulation Methods in Statistical Physics, Project Report, FK7029, Stockholm Universitysebastian100% (2)
- HT TP: //qpa Pe R.W But .Ac .In: Data Compression & CryptographyDocument7 pagesHT TP: //qpa Pe R.W But .Ac .In: Data Compression & CryptographyDuma DumaiNo ratings yet
- Count Trailing Zeroes in Factorial of A Number - GeeksforGeeksDocument13 pagesCount Trailing Zeroes in Factorial of A Number - GeeksforGeeksWatan SahuNo ratings yet
- Question of Chapter 8 StaticsDocument7 pagesQuestion of Chapter 8 Staticsحمزہ قاسم کاھلوں جٹNo ratings yet
- Intro Bridge ProjectDocument33 pagesIntro Bridge ProjectTarak A PositiveNo ratings yet
- (Springer Series in Materials Science 200) Ilya V. Shadrivov, Mikhail Lapine, Yuri S. Kivshar (Eds.) - Nonlinear, Tunable and Active Metamaterials-Springer International Publishing (2015) PDFDocument333 pages(Springer Series in Materials Science 200) Ilya V. Shadrivov, Mikhail Lapine, Yuri S. Kivshar (Eds.) - Nonlinear, Tunable and Active Metamaterials-Springer International Publishing (2015) PDFDebidas KunduNo ratings yet
- Series Quesions For IQDocument2 pagesSeries Quesions For IQkanaya khanNo ratings yet
- This Study Resource WasDocument4 pagesThis Study Resource WasYash GargNo ratings yet
- 동역학12 PDFDocument13 pages동역학12 PDF엄석현No ratings yet
- Lecture 1 CONFIDENCE INTERVALDocument49 pagesLecture 1 CONFIDENCE INTERVALEmmmanuel ArthurNo ratings yet
- Math 7 ADM Module 7 EditedDocument26 pagesMath 7 ADM Module 7 EditedMyrna Domingo Ramos0% (1)
- Factoring PolynomialsDocument3 pagesFactoring PolynomialsdawnbrodieNo ratings yet
- Fir Kaiser Window Notes-3 PDFDocument19 pagesFir Kaiser Window Notes-3 PDFprajwalNo ratings yet
- Foundation Faculty Recruitment Test 2018 - (Code-A) - MAbilityDocument5 pagesFoundation Faculty Recruitment Test 2018 - (Code-A) - MAbilityjitendra bhardwajNo ratings yet
- Acme ThreadsDocument5 pagesAcme ThreadsBob WarfieldNo ratings yet