Professional Documents
Culture Documents
Ni Hms 23469
Ni Hms 23469
Uploaded by
Naresh Prasad SapkotaCopyright:
Available Formats
You might also like
- Molecular Biology of The Cell, 5th EditionDocument82 pagesMolecular Biology of The Cell, 5th EditionBee Nunes25% (67)
- Active Reading: Answer KeyDocument3 pagesActive Reading: Answer KeyJonas Montero100% (2)
- Bio Calculus PDocument738 pagesBio Calculus PMarco Sousa100% (1)
- Molecular Population Genetics and Evolution - Masatoshi NeiDocument290 pagesMolecular Population Genetics and Evolution - Masatoshi NeiGeorge Sebastian Antony100% (3)
- Extended Heredity: A New Understanding of Inheritance and EvolutionFrom EverandExtended Heredity: A New Understanding of Inheritance and EvolutionNo ratings yet
- PHYS 444 - Physical Biology: From Molecules To Cells Fall 2014 Course InformationDocument6 pagesPHYS 444 - Physical Biology: From Molecules To Cells Fall 2014 Course InformationAnonymous bZTdTpLNo ratings yet
- Coursework B Science 2014 BiologyDocument4 pagesCoursework B Science 2014 Biologyafiwjsazh100% (2)
- Cbe 513Document11 pagesCbe 513duverney.gaviriaNo ratings yet
- Applications of Mathematics To Biology: Manuscript ID: #0443 Original Research PaperDocument5 pagesApplications of Mathematics To Biology: Manuscript ID: #0443 Original Research Paperrizka arzitaNo ratings yet
- The Crossroads Between Biology and Mathematics: The Scientific Method As The Basics of Scientific LiteracyDocument8 pagesThe Crossroads Between Biology and Mathematics: The Scientific Method As The Basics of Scientific LiteracySantiago OrtegonNo ratings yet
- Thesis On Evolutionary BiologyDocument4 pagesThesis On Evolutionary Biologydwfz48q3100% (2)
- A Neural-Functionalist Approach To Learning: Texas A&M University, U.S.ADocument22 pagesA Neural-Functionalist Approach To Learning: Texas A&M University, U.S.AAna HernandezNo ratings yet
- Literature Review Molecular BiologyDocument7 pagesLiterature Review Molecular Biologyafdtsebxc100% (1)
- Misconception About Genetic DriftDocument12 pagesMisconception About Genetic DriftNathalia LaseNo ratings yet
- Admin,+10994 Kloser1Document18 pagesAdmin,+10994 Kloser1Tiago GuimarãesNo ratings yet
- Literary Practices of Biology 1Document6 pagesLiterary Practices of Biology 1api-361092487No ratings yet
- Multiple Representations in Biological Education 2013Document394 pagesMultiple Representations in Biological Education 2013Miguel CalderonNo ratings yet
- Essay 1Document7 pagesEssay 1api-361089929No ratings yet
- Science2004-May-Math in BioDocument5 pagesScience2004-May-Math in BioÁfrica RamosNo ratings yet
- Biophysicsconcep00case PDFDocument360 pagesBiophysicsconcep00case PDFaabc100% (1)
- Biophysics Conce P 00 CaseDocument360 pagesBiophysics Conce P 00 Casemagini3649No ratings yet
- Human Biology Topics For Research PapersDocument7 pagesHuman Biology Topics For Research Papersgz9g97ha100% (1)
- Odom Diffusion Osmosis VeryDocument27 pagesOdom Diffusion Osmosis VerycelestinedordorNo ratings yet
- Leszek Konieczny, Irena Roterman-Konieczna, Paweł Spólnik - Systems Biology - Functional Strategies of Living Organisms-Springer (2023) (Z-Lib - Io)Document269 pagesLeszek Konieczny, Irena Roterman-Konieczna, Paweł Spólnik - Systems Biology - Functional Strategies of Living Organisms-Springer (2023) (Z-Lib - Io)Garbage CanNo ratings yet
- Biology Senior Thesis ExamplesDocument8 pagesBiology Senior Thesis Examplesstephaniebenjaminclarksville100% (1)
- Biology and MathematicsDocument7 pagesBiology and MathematicsMaria AnghelacheNo ratings yet
- Mathematical Manipulative Models: in Defense of "Beanbag Biology"Document11 pagesMathematical Manipulative Models: in Defense of "Beanbag Biology"jungck8715No ratings yet
- Garcia2007b PDFDocument39 pagesGarcia2007b PDFMarciNo ratings yet
- The Basics of Biology - C. StoneDocument294 pagesThe Basics of Biology - C. Stonemonicq77100% (1)
- Bio2010, Report in BriefDocument8 pagesBio2010, Report in BriefNational Academies of Science, Engineering, and MedicineNo ratings yet
- Goriely 2017Document652 pagesGoriely 2017Тимофей НестеровNo ratings yet
- Demographic Methods Across The Tree of Life Roberto Salguero Gomez Full ChapterDocument66 pagesDemographic Methods Across The Tree of Life Roberto Salguero Gomez Full Chapterwilliam.belliveau809100% (21)
- SafariDocument3 pagesSafarinerinajonasNo ratings yet
- Chemistry Lessons For Universities A Review of Constructivist IDEASDocument10 pagesChemistry Lessons For Universities A Review of Constructivist IDEASVictor Manuel Enriquez GNo ratings yet
- Progress in Theoretical Biology: Volume 1From EverandProgress in Theoretical Biology: Volume 1Fred M. SnellNo ratings yet
- Evolution in Changing Environments: Some Theoretical Explorations. (MPB-2)From EverandEvolution in Changing Environments: Some Theoretical Explorations. (MPB-2)No ratings yet
- Molecular Biology Dissertation IdeasDocument4 pagesMolecular Biology Dissertation IdeasBuyCollegePaperOnlineUK100% (1)
- Making Sense of Evolution: The Conceptual Foundations of Evolutionary BiologyFrom EverandMaking Sense of Evolution: The Conceptual Foundations of Evolutionary BiologyRating: 4 out of 5 stars4/5 (3)
- Biology Education Thesis PDFDocument5 pagesBiology Education Thesis PDFPaperHelpWritingCanada100% (1)
- Science ReflectionDocument2 pagesScience Reflectionapi-281124194100% (1)
- Human Genetics by SCDocument626 pagesHuman Genetics by SCtaniyaNo ratings yet
- Uses and Abuses of Mathematics in BiologyDocument4 pagesUses and Abuses of Mathematics in Biologymuhammad sami ullah khanNo ratings yet
- Biologia Matemática 2017Document20 pagesBiologia Matemática 2017iago.sk66No ratings yet
- How To Write A Biology Literature ReviewDocument7 pagesHow To Write A Biology Literature Reviewaiqbzprif100% (1)
- Thesis MicrobiologyDocument8 pagesThesis Microbiologywyppfyhef100% (2)
- The University of Chicago Press Philosophy of Science AssociationDocument11 pagesThe University of Chicago Press Philosophy of Science AssociationAbuAbdur-RazzaqAl-MisriNo ratings yet
- Introductory Physics For The Life Sciences by Simon MochrieDocument860 pagesIntroductory Physics For The Life Sciences by Simon MochrieAina Horrach AlonsoNo ratings yet
- Alberts Introductory PagesDocument33 pagesAlberts Introductory PagesMarianne GonzalesNo ratings yet
- Hugh Drummond (Auth.), Paul Patrick Gordon Bateson, Peter H. Klopfer (Eds.) - Perspectives in Ethology - Volume 4 Advantages of Diversity (1981, Springer US)Document253 pagesHugh Drummond (Auth.), Paul Patrick Gordon Bateson, Peter H. Klopfer (Eds.) - Perspectives in Ethology - Volume 4 Advantages of Diversity (1981, Springer US)Mauro AraújoNo ratings yet
- Biology Literature ReviewDocument4 pagesBiology Literature Reviewjzczyfvkg100% (1)
- Active Voice in ScienceDocument2 pagesActive Voice in SciencelalaNo ratings yet
- Teaching Evolution & The Nature of Science Farber 2003Document9 pagesTeaching Evolution & The Nature of Science Farber 2003Chrystal CarattaNo ratings yet
- Thesis ZoologyDocument7 pagesThesis Zoologytsfublhld100% (2)
- Information and Living Systems BOOKDocument459 pagesInformation and Living Systems BOOKeduardoazevedo_ccciNo ratings yet
- Biology's Next RevolutionDocument1 pageBiology's Next RevolutionbarrospiresNo ratings yet
- Exploring The Limitations of The Scientific MethodDocument4 pagesExploring The Limitations of The Scientific MethodADNo ratings yet
- Introduction To The Philosophy of Science First PartDocument138 pagesIntroduction To The Philosophy of Science First PartChandra Sekhar Sahu100% (1)
- Coek - Info Molecular Biology 2nd EdnDocument1 pageCoek - Info Molecular Biology 2nd EdniqrarNo ratings yet
- Modeling in Computational Biology and Biomedicine:: A Multidisciplinary Endeavor Draft (April 2012)Document31 pagesModeling in Computational Biology and Biomedicine:: A Multidisciplinary Endeavor Draft (April 2012)Dessi DuckNo ratings yet
- Seafood Production - Our World in DataDocument14 pagesSeafood Production - Our World in DataNaresh Prasad SapkotaNo ratings yet
- LGW (G Ptyfg N3'Ljq Ljqlo +:yf Lnld6) 8Sf) T) - F) Q) DFDocument7 pagesLGW (G Ptyfg N3'Ljq Ljqlo +:yf Lnld6) 8Sf) T) - F) Q) DFNaresh Prasad SapkotaNo ratings yet
- My Philosophy - Greg MatDocument5 pagesMy Philosophy - Greg MatNaresh Prasad SapkotaNo ratings yet
- 2019年01月27日GRE考试真题Document22 pages2019年01月27日GRE考试真题Naresh Prasad SapkotaNo ratings yet
- Happy Painting GuideBookDocument3 pagesHappy Painting GuideBookNaresh Prasad SapkotaNo ratings yet
- Bachelor Curriculum Bioinformatics: (In English)Document12 pagesBachelor Curriculum Bioinformatics: (In English)Naresh Prasad SapkotaNo ratings yet
- Week Two Lesson Summaries - CourseraDocument6 pagesWeek Two Lesson Summaries - CourseraNaresh Prasad SapkotaNo ratings yet
- Manjushree Finance - Stock AnalysisDocument9 pagesManjushree Finance - Stock AnalysisNaresh Prasad SapkotaNo ratings yet
- Back To Week 2 LessonsDocument5 pagesBack To Week 2 LessonsNaresh Prasad SapkotaNo ratings yet
- NEWGREPrepByMagoosh 2017Document469 pagesNEWGREPrepByMagoosh 2017Techo Sunny100% (25)
- Dictyostelium Amoebae Are Free Living Cells With A Remarkable Twist: Under The Stress of StarvationDocument4 pagesDictyostelium Amoebae Are Free Living Cells With A Remarkable Twist: Under The Stress of StarvationNaresh Prasad SapkotaNo ratings yet
- L14: 14 Lectures Past and 11 To Go Part I: The Cell As A Well-Stirred Bioreactor'Document3 pagesL14: 14 Lectures Past and 11 To Go Part I: The Cell As A Well-Stirred Bioreactor'Naresh Prasad SapkotaNo ratings yet
- Fine Tuned Model For Perfect Adaptation'Document14 pagesFine Tuned Model For Perfect Adaptation'Naresh Prasad SapkotaNo ratings yet
- Creation of Membrane Associated MindDocument4 pagesCreation of Membrane Associated MindNaresh Prasad SapkotaNo ratings yet
- Engineering Biology To Do Chemistry: BE.010 Spring 2005 Session # 3 NotesDocument2 pagesEngineering Biology To Do Chemistry: BE.010 Spring 2005 Session # 3 NotesNaresh Prasad SapkotaNo ratings yet
- Goals:: Tips To Writing A Successful Proposal For 6.021JDocument2 pagesGoals:: Tips To Writing A Successful Proposal For 6.021JNaresh Prasad SapkotaNo ratings yet
- D Re D: Waiting Times For Chemical ReactionsDocument2 pagesD Re D: Waiting Times For Chemical ReactionsNaresh Prasad SapkotaNo ratings yet
- Summary Lecture 3: Image by MIT OCWDocument6 pagesSummary Lecture 3: Image by MIT OCWNaresh Prasad SapkotaNo ratings yet
- Ps 1Document2 pagesPs 1Naresh Prasad SapkotaNo ratings yet
- Chia-Pet (Chromatin Interaction Analysis by Paired-End Tag Sequencing)Document13 pagesChia-Pet (Chromatin Interaction Analysis by Paired-End Tag Sequencing)Naresh Prasad SapkotaNo ratings yet
- Slides Module01 UsefulGeneticsLecture1F-SlidesDocument13 pagesSlides Module01 UsefulGeneticsLecture1F-SlidesNaresh Prasad SapkotaNo ratings yet
- l16 NotesDocument10 pagesl16 NotesNaresh Prasad SapkotaNo ratings yet
- Systems Biology: X Y X YDocument2 pagesSystems Biology: X Y X YNaresh Prasad SapkotaNo ratings yet
- I Systems Microbiology (13 Lectures) : The Cell As A Well-Stirred Biochemical Reactor'Document20 pagesI Systems Microbiology (13 Lectures) : The Cell As A Well-Stirred Biochemical Reactor'Naresh Prasad SapkotaNo ratings yet
- Final ProjectDocument14 pagesFinal ProjectShivangi VaidNo ratings yet
- LacusCurtius - Strabo's Geography - Book III Chapter 3Document10 pagesLacusCurtius - Strabo's Geography - Book III Chapter 3A Miguel Simão LealNo ratings yet
- Reading and Writing TOS With ExamDocument2 pagesReading and Writing TOS With Examjoy grace jaraNo ratings yet
- MEETING 5 - Describing PeopleDocument6 pagesMEETING 5 - Describing Peoplerizky.ameliaNo ratings yet
- Team EkowDocument4 pagesTeam EkowDavid Ekow NketsiahNo ratings yet
- Banner Value BeamDocument20 pagesBanner Value BeamryanmackintoshNo ratings yet
- OsiSense XU XUK8TAE1MM12 DocumentDocument6 pagesOsiSense XU XUK8TAE1MM12 Documentحمزة صباحNo ratings yet
- Journal of Pharmaceutical Health Services ResearchDocument2 pagesJournal of Pharmaceutical Health Services ResearchTrần Văn ĐệNo ratings yet
- TR 6Document16 pagesTR 6Hritika RajvanshiNo ratings yet
- Erie Scientific LLC: Globally Harmonized Safety Data Sheet (GHSDS)Document7 pagesErie Scientific LLC: Globally Harmonized Safety Data Sheet (GHSDS)Leni MeifitaNo ratings yet
- SilverCrest SHFD 1350 A1 Deep Fryer AppleDocument128 pagesSilverCrest SHFD 1350 A1 Deep Fryer AppleberjanatNo ratings yet
- Duraplate 8200Document4 pagesDuraplate 8200Carlos VillacrizNo ratings yet
- Topographic Map of Flag PondDocument1 pageTopographic Map of Flag PondHistoricalMapsNo ratings yet
- PA4600 & PA6000 User Manual - Rev GDocument53 pagesPA4600 & PA6000 User Manual - Rev GjatzireNo ratings yet
- OSCP Personal Cheatsheet: September 18Th, 2020 o o o oDocument76 pagesOSCP Personal Cheatsheet: September 18Th, 2020 o o o omahesh yadavNo ratings yet
- Organizational Behavior Human Behavior at Work 14th Edition Newstrom Test BankDocument16 pagesOrganizational Behavior Human Behavior at Work 14th Edition Newstrom Test Bankzaneraymondjpanf100% (28)
- Hit and Miss Brickwork: BDA Design Note 02Document2 pagesHit and Miss Brickwork: BDA Design Note 02SamNo ratings yet
- Asphalt Surfacing To Bridge DecksDocument44 pagesAsphalt Surfacing To Bridge DecksNguyễn Văn MinhNo ratings yet
- Simulated Binar Crossover F Continuous Search Space: Comp SystemsDocument34 pagesSimulated Binar Crossover F Continuous Search Space: Comp SystemsSal Ie EmNo ratings yet
- DC Motors Introduction, Equivalent Circuit, TypesDocument22 pagesDC Motors Introduction, Equivalent Circuit, Typesmalik karim dadNo ratings yet
- ConditionalsDocument16 pagesConditionalsAdalid Pulido AriasNo ratings yet
- Statistics Final Pre TestDocument4 pagesStatistics Final Pre TestLorienelNo ratings yet
- It Cleft PDFDocument280 pagesIt Cleft PDFmeannaNo ratings yet
- 593c0be3230b5002a4a21249 Daniel Guillot Ortiz PDFDocument80 pages593c0be3230b5002a4a21249 Daniel Guillot Ortiz PDFElisa MartinezNo ratings yet
- OmniOMS Output GenerationDocument2 pagesOmniOMS Output Generationnitin.aktivNo ratings yet
- Bombardier Data SheetDocument4 pagesBombardier Data SheetLalu Giat AjahNo ratings yet
- Sub-Project Proposal: Municipality of Kabayan Barangay TAWANGANDocument6 pagesSub-Project Proposal: Municipality of Kabayan Barangay TAWANGANpres carap60% (5)
- MTU 10V1600 DS450: Diesel Generator SetDocument4 pagesMTU 10V1600 DS450: Diesel Generator SetJorge Eraldo Albarran PoleoNo ratings yet
- Fin TechDocument32 pagesFin Techkritigupta.may1999No ratings yet
Ni Hms 23469
Ni Hms 23469
Uploaded by
Naresh Prasad SapkotaCopyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Ni Hms 23469
Ni Hms 23469
Uploaded by
Naresh Prasad SapkotaCopyright:
Available Formats
NIH Public Access
Author Manuscript
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
NIH-PA Author Manuscript
Published in final edited form as:
Nat Rev Mol Cell Biol. 2006 November ; 7(11): 829832.
Back to the future: education for systems-level biologists
Ned Wingreen and David Botstein
Ned Wingreen is at the Department of Molecular Biology, and David Botstein is at the Lewis-Sigler
Institute and the Department of Molecular Biology, Princeton University, Princeton, New Jersey
08544, USA.
Abstract
We describe a graduate course in quantitative biology that is based on original path-breaking papers
in diverse areas of biology; each of these papers depends on quantitative reasoning and theory as
well as experiment. Close reading and discussion of these papers allows students with backgrounds
in physics, computational sciences or biology to learn essential ideas and to communicate in the
languages of disciplines other than their own.
NIH-PA Author Manuscript
The genome has presented biologists with an opportunity to study genetic processes on a
genomic scale, and to achieve quantitative understanding, not just of individual molecular
mechanisms but also of their interactions and regulation at the systems level. The
transformation of biology into a fully quantitative, theory-rich science now seems inevitable,
if not yet quite within reach.
These developments have produced a challenge to the educational system. It is clear that the
future of biology will require combinations of skills that are rarely found in individual scientists
today. The existing educational system teaches biologists very few mathematical or
computational skills, and gives scientists with backgrounds in physics and informatics
comparably limited exposure to even the most basic biological phenomena and principles. The
problem begins early in undergraduate education, and by the doctoral level there are severe
interdisciplinary communication difficulties that are encountered by even the most motivated
of collaborators1.
An integrated science curriculum
NIH-PA Author Manuscript
At Princeton, we have begun to address the educational challenges at both the undergraduate
and the graduate levels. Our goal, at each level, is to equip students to succeed across scientific
disciplines using the language and mathematics appropriate to each. To this end, we are
teaching courses that integrate subjects that were traditionally taught separately, such as
mathematics, physics, chemistry and biology. We are finding that, at the undergraduate level,
integration works best when it is initiated in the first year of college. An integrated science
curriculum, taught at the level of the most challenging physics, computation, chemistry, and
biology courses, serves to bring the students to a level of sophistication that allows them to
major in any of the sciences.
Correspondence to D.B. e-mail: botstein@princeton.edu
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Ned Wingreenss homepage: http://www.molbio.princeton.edu/research_facultymember.php?id=65. David Botsteins homepage:
http://www.molbio.princeton.edu/research_facultymember.php?id=60
Access to this links box is available online.
Wingreen and Botstein
Page 2
NIH-PA Author Manuscript
At the graduate level the problem is more difficult as the students have already differentiated
to some extent and view themselves as, for example, physicists, chemists or biologists. We
find that first-year graduate students already have some lacunae in their education: the
biologists have had limited education in physics, computation and mathematics, and the
physical and computational scientists have little or no biology at their command. The
temptation is to prescribe remedial undergraduate courses; however, this approach does not
appeal to graduate students. Our alternative is to begin with an integrated graduate course in
quantitative biology.
We teach a single course to a population that consists of approximately equal numbers of
graduate students in biology and physics, mostly in their first or second years. These students
are interested in the interface between biology and quantitative science (FIG. 1). We meet with
the students together, in a seminar format, having assigned two papers often classics that
use sophisticated quantitative methods and concepts to study biological problems. We discuss
the papers in detail, but not necessarily in the original order of presentation, for almost three
hours. We use, with some success, the skills of one group of students to help the other group;
when biological issues arise, the biologists tend to speak, and when quantitative issues are on
the table, the physicists tend to participate most actively. Each group develops increasing
respect for the value of listening to the other, and we are optimistic that the students become
motivated to help each other.
NIH-PA Author Manuscript
Course materials
The nature and quality of the papers is crucial to the success of this course. The papers need
to be sufficiently sophisticated and important to repay detailed study. The papers also have to
function as vehicles for teaching both biology and quantitative analysis. We have found,
somewhat to our surprise, that some of the best papers are quite old, possibly harking back to
a time when biologists and physical scientists educations were less different than they are
today. We have also found that the lessons in these old papers are being re-emphasized in the
current systems biology literature (BOX 1).
In this section, we present nearly a dozen of the pedagogically most successful papers. We also
provide a few words on the lessons we find embodied in them.
Random processes and distributions
Luria, S.E. and Delbrck, M. Mutations of bacteria from virus sensitivity to virus resistance
(1943)2.
NIH-PA Author Manuscript
This classic paper, which was published more than 50 years ago, first presented the appropriate
mathematics for analysing random mutations as they arise in a population. This mathematical
analysis can be applied to a class of problems in biology that, in subsequent years, has been
shown to also involve statistical physics and chemistry. The analysis begins with an
understanding of Poisson processes and the Poisson distribution (see Glossary).
Luria and Delbrck addressed the specific problem of mutations arising in a growing colony
of Escherichia coli cells derived from a single phage-sensitive ancestor. To analyse their data
they had to consider the distribution of cells that have mutated to resistance to bacteriophage
attack. The LuriaDelbrck distribution has a remarkably long tail, due to the occasional
jackpot in which a resistance-endowing mutation occurred at an early generation in the history
of the clone. It was, indeed, the use of a slot machine that stimulated Luria to think about
mutation in this way, and the entirely appropriate term jackpot was used by him.
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 3
NIH-PA Author Manuscript
As well as teaching the students to think about random processes in populations (for homework,
see BOX 2), this paper introduces a considerable amount of basic microbiology. A good
example lies in the primary motivation for the study. Does resistance to phage arise from
random mutations, or from the development of immunity after exposure? We ask the students
to discuss questions such as: what if, instead of studying the lytic phage T1, Luria and Delbrck
had studied the lysogenic phage , which can integrate into the genome and provide immunity?
Individuality
Elowitz, M.B. et al. Stochastic gene expression in a single cell (2002)3.
This is a very good paper for discussing the individuality of genetically identical cells, which
is rapidly becoming the standard paradigm for studying stochastic gene expression in single
cells. Simultaneous measurement of two identically regulated fluorescent proteins in single
cells distinguishes intrinsic, biochemical noise from extrinsic, cell-to-cell variation. Of course,
what gets classed as extrinsic and intrinsic depends on how the two reporters are constructed;
for example, if the reporters are on opposite sides of the chromosome from the origin of
replication, what happens in strains in which replication can stall?
NIH-PA Author Manuscript
This paper is useful for discussing how noise propagates from DNA copies, to mRNA copies,
to number of proteins. More generally, it teaches the importance of single-cell measurements;
what does one fail to notice in population-averaged measurements?
Stable switching
Novick, A. and Wiener, M. Enzyme induction as an all-or-none phenomenon (1957)4.
This is another classic paper, not nearly so well known today as the Luria and Delbrck paper,
but equally influential in its time. It provides a complete and convincing analysis of how
genetically identical cells can stably maintain two heritable states. The lac operon, as an
experimental system, remains the premier model not only for classic gene-expression
regulation, but also for systems-level analysis (BOX 1).
Over a range of lactose concentrations, the lac system supports two stable states: induced and
uninduced. The papers most interesting lesson is that the history of the cells matters. Another
basic biology (as well as mathematical) lesson is that the presence or absence of permease
molecules determines the nature of diauxic growth that is, permease molecules are the agents
of a cells memory in the lac system. The comparison of growth rates of induced and
uninduced cells foreshadows the recent interest in quantifying fitness (see also REF. 5).
NIH-PA Author Manuscript
Robustness
Barkai, N. and Leibler, S. Robustness in simple biochemical networks (1997)6.
An elegant example of how robustness preservation of function despite fluctuations in
components can be built-in at the network level. Individual E. coli cells adapt precisely to
ambient levels of chemo-attractants, in some cases over a range of five orders of magnitude,
despite fluctuations of network components.
Barkai and Leibler proposed a model in which active and inactive receptors are distinguished
by different net rates of methylation. To achieve steady state, the average methylation and
demethylation rates must come into balance, which only happens when receptors reach a
particular average activity, therefore ensuring the precise adaptation of receptor activity. The
wealth of information about the chemotaxis network makes this system the model for
understanding signal transduction at a quantitative level.
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 4
Ultrasensitivity
NIH-PA Author Manuscript
Goldbeter, A. and Koshland, D.E. Jr. An amplified sensitivity arising from covalent
modification in biological systems (1981)7.
Chemical modification of proteins by enzymatic cycles (for example, phosphorylation and
dephosphorylation) lies at the heart of many cellular signalling systems. Goldbeter and
Koshland analysed the inputoutput characteristics of such cycles. How do changes in enzyme
levels affect the steady-state level of a modified protein? Increasing enzyme saturation results
in increasing ultrasensitivity that is, a sharper, graphically more-step-like response. The
paper segues smoothly between mathematics and biology, and provides critical insights into
our current understanding of developmental switches in cells.
Specificity
Hopfield, J.J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic
processes requiring high specificity (1974)8.
NIH-PA Author Manuscript
How do cells achieve specificity? In a classic paper that is more relevant today than ever,
Hopfield showed that a simple, kinetic (energy-driven) mechanism can multiplicatively
enhance the intrinsic specificity of enzymesubstrate reactions. Examples of kinetic
proofreading are taken from transcription and translation, and the mechanism beautifully
explains the seemingly wasteful, extra energy inputs to these processes. This paper teaches that
when studying biological processes, one should ask; what might go wrong and at what rate
would such errors be expected? When there is a mismatch between expectation and
observation, as there was in the fidelity of protein synthesis in the decade before Hopfields
paper, then new biological processes might be waiting to be discovered.
Similarity
Smith, T.F. and Waterman, M.S. Identification of common molecular subsequences (1981)9.
Smith and Waterman tackled the problem of local sequence alignment. This seminal paper
provides students with a good introduction to the problem, and an appreciation for the
importance of how different algorithms scale with problem size. How does the use of dynamic
programming bypass the (exponentially difficult) task of comparing every possible alignment
of two sequences? This study also provides a springboard for discussing what one means by
similarity, and how homology (evolution from a common ancestor) influences our
understanding of the meaning of statistically unlikely similarities between sequences.
NIH-PA Author Manuscript
Maximum likelihood
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach
(1981)10.
How do we distinguish between different models given a particular set of data? Felsenstein
presents a simple and clearly written example using alternative unrooted phylogenetic trees as
models and DNA sequences from extant species as data. Given a simple assumption for
mutation rates, which tree is most likely to lead to the observed data? How confident can we
be in the maximum likelihood solution? This article is a good lead-in to a discussion of Bayesian
analysis and its wide application to large, and potentially mixed, data sets in this era of genomics
and systems-level biology.
Evolutionary perspective
Eisen, J.A. A phylogenomic study of the MutS family of proteins (1998)11.
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 5
NIH-PA Author Manuscript
Another important aspect of the course is to introduce the idea of phylogenomics knowledge
of genomes is vital to determining the evolutionary tree, and knowledge of the evolutionary
tree is a powerful guide to interpreting genomes. Eisen presents a particularly good example:
the MutS family of DNA-repair proteins that is found in virtually all organisms. The function
of MutS has changed in several ways along different lineages, which becomes readily apparent
only when studying the evolutionary tree.
Microarray-data analysis
Eisen, M.B. et al. Cluster analysis and display of genome-wide expression patterns (1998)
12.
The analysis of microarray data using correlation coefficients between genes and hierarchical
clustering has stood the test of time, compared to more complex approaches. This paper is also
a good introduction to systems-level thinking about cellular dynamics. It is apparent from the
data that genes dont turn on and off one at a time, rather, cohorts of genes turn on and off
together. We stress to the students the importance of data visualization. The similarity of geneexpression patterns can be seen at a glance when data are presented as coloured patterns. We
ask the students to imagine the same data presented as the table of numbers that underlie these
coloured patterns. We also contrast this kind of analysis with methods that require removing,
summarizing or otherwise changing the underlying data.
NIH-PA Author Manuscript
Biophysical modeling
Hodgkin, A.L. Croonian Lecture, ionic movements and electrical activity in giant nerve fibres
(1958)13.
An important theme in quantitative biology is biophysical modelling. This lecture presents a
beautiful example of biophysical modelling that is the foundation of neurobiology. The story
of Hodgkin and Huxleys study of giant nerve fibres runs step-by-step from the basic biology
and physiology of axons and action potentials, through the crucial electrophysiology
experiments, to the now famous HodgkinHuxley equations and their solution.
The students learn something about electrical engineering, ion channels, chemical potentials
and differential equations, and a lot about reasoning from experiment. An alternative to this
Croonian Lecture, which we have considered but not tried, might be to use one or more of the
original Hodgkin and Huxley publications, as we think that papers presenting original data are
probably more effective in such a course than reviews.
NIH-PA Author Manuscript
Conclusions
To summarize our experience, we have found that close reading of papers that made a
significant contribution to basic biology through the use of quantitative reasoning and theory
is useful in the setting of first-year graduate students with diverse educational backgrounds.
The papers help those students with a biology background to focus on the mathematical and
computational ideas and methods that are most relevant not only to current practice, but also
to the students areas of interest.
In a complementary way, the students with physics and computational backgrounds are
exposed to many issues of significance in basic biology. They also become familiar with
research areas to which they might well be able to contribute, if they continue to pursue their
interests in biology. Last, we are hopeful that the emerging practice of teaching students with
such diverse backgrounds together will facilitate communication and professional relationships
that will serve these future members of the genomic and systems biology community well.
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 6
Box 1 Diauxie understanding a paradigm in gene regulation
NIH-PA Author Manuscript
This figure, from Jacques Monods Ph.D. dissertation (La Croissance des Cultures
Bactriennes, 1942), illustrates a basic phenomenon that led directly to the discovery of
gene regulation by repressors in bacteria, and to a Nobel Prize. This figure (or something
similar to it, usually with lactose instead of galactose as the second sugar) is in virtually
every textbook of genetics and molecular or cell biology.
However, understanding of this phenomenon is not possible without a quantitative analysis,
which is introduced in one of the papers read in our course7. The persistent failure of the
galactose (or lactose) operon to be induced in the presence of inducer depends crucially on
the absence of permease molecules. Novick and Weiner7 clearly showed that diauxie works
because induction is a positive-feedback system in which the inducer is taken up from the
medium by a permease molecule that is itself induced. It is this feature that makes the
relatively weak phenomenon of catabolite repression into an absolute barrier to induction.
Because this phenomenon illustrates so nicely the importance of feedback and memory, it
is still studied today by systems-level biologists (see REF 14)
NIH-PA Author Manuscript
Box 2 A representative homework problem: Beyond grad school!
NIH-PA Author Manuscript
Problem. Youre the technical advisor for a group of investors that are interested in buying
a company that engineers bacteria to detoxify waste. An important issue is that the
engineered bacteria are slightly less fit than the wild-type in the wild (where they will be
used) but not in the laboratory (where they will be grown up and packaged for sale dispersal).
The engineered bacteria revert (mutate) to wild-type in the laboratory at a rate that is,
at each cell division there is a probability that one daughter will become wild-type, and
then that all of the daughters progeny remain wild-type. The company has performed a set
of 10 experiments each starts with a single engineered cell, which grows for 20
generations, and then measures the number of wild-type (revertant) cells. In these 10 trials,
they found 0, 0, 8, 14, 16, 24, 38, 44, 70 and 80 revertant cells. The company statisticians
argue that the probability of getting 0 revertants is given by the probability of the 0 outcome
in a Poisson process, namely p (0) = e, in which the mean is given by the expected number
of mutation events in a colony, that is, = 220. So, taking the probability of 0 revertants
from the data to be 2/10, they estimate that = 1.61, which corresponds to = 1.5 106.
The investors have asked you to verify this estimate for the reversion rate. They are only
interested in buying the company if the rate is below 2 106. Your intuition is that you
can get a more reliable estimate for the rate using the entire data set rather than just the
fraction of trials with 0 revertants.
Use the maximum likelihood approach to find the mutation rate for which the observed
data has the highest likelihood (you will need to generate accurate distributions of revertants
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 7
for different values of, calculate the likelihood of the observed data for each, and find
the that maximizes this likelihood). Should the investors buy the company?
NIH-PA Author Manuscript
Authors note. The intention of this problem was to have the students do an exhaustive
simulation, a technique not available to Delbrck in 1943. However, when this problem
was given, several of the students realized that in addition to the possibility of simulating
the full LuriaDelbrck distribution, it is possible to analytically calculate the distribution
for the low numbers used here. We thought this was a great success.
The paper provides a good introduction to the diversity of evolutionary events that must be
considered, including gene duplications, gene deletions, horizontal transfer and, of course,
speciation. More generally, it shows the value of taking an evolutionary perspective in
interpreting biological function.
References
NIH-PA Author Manuscript
1. Bialek W, Botstein D. Introductory science and mathematics education for 21st century biologists.
Science 2004;303:788 790. [PubMed: 14764865]
2. Luria SE, Delbrck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics
1943;28:491511. [PubMed: 17247100]
3. Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science
2002;297:11831186. [PubMed: 12183631]
4. Novick A, Wiener M. Enzyme Induction as an all-or-none phenomenon. Proc Natl Acad Sci USA
1957;43:553566. [PubMed: 16590055]
5. Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature
2005;436:588592. [PubMed: 16049495]
6. Barkai N, Leibler S. Robustness in simple biochemical networks. Nature 1997;387:913917. [PubMed:
9202124]
7. Goldbeter A, Koshland DE Jr. An amplified sensitivity arising from covalent modification in biological
systems. Proc Natl Acad Sci USA 1981;78:68406844. [PubMed: 6947258]
8. Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes
requiring high specificity. Proc Natl Acad Sci USA 1974;71:41354139. [PubMed: 4530290]
9. Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol
1981;147:195197. [PubMed: 7265238]
10. Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol
1981;17:368376. [PubMed: 7288891]
11. Eisen JA. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res 1998;26:4291
4300. [PubMed: 9722651]
12. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide
expression patterns. Proc Natl Acad Sci USA 1998;95:1486314868. [PubMed: 9843981]
13. Hodgkin AL. Croonian Lecture, ionic movements and electrical activity in giant nerve fibres. Proc
R Soc Lond B Biol Sci 1958;148:137. [PubMed: 13494473]
14. Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose
utilization network of Escherichia coli. Nature 2004;427:737740. [PubMed: 14973486]
NIH-PA Author Manuscript
Glossary
Croonian Lecture
The Croonian Lectures are prestigious lectureships given at the invitation of the
Royal Society and the Royal College of Physicians.
Diauxic growth
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 8
NIH-PA Author Manuscript
In a medium that contains glucose and a less preferred carbon source, bacteria
exhaust the glucose before consuming the other carbon source. Monod called this
behaviour diauxie (double growth in French).
HodgkinHuxley equations
Hodgkin and Huxley published a series of (now classic) papers in 1952 on
electrical activity and transmembrane ion currents in the squid giant axon. In
these papers, they derived the HodgkinHuxley equations, which accurately
describe the action potential.
Jackpot
In a series of parallel cultures, the LuriaDelbrck jackpot is the rare
observation of a large clone of mutant cells that are derived from a single
mutational event early in the growth of the culture. Though rare, these jackpots
occur much more frequently than the probability p(n) that is expected from simple
Poisson statistics.
Poisson distribution
This distribution, p(n) = exp() n/n!, gives the probability of observing n rare
random events in a very large population, for which is the average expected
number of these rare events.
NIH-PA Author Manuscript
NIH-PA Author Manuscript
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
Wingreen and Botstein
Page 9
NIH-PA Author Manuscript
NIH-PA Author Manuscript
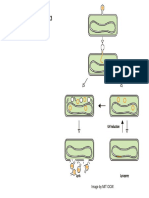
Figure 1.
Taking integrated systems biology a step too far.
NIH-PA Author Manuscript
Nat Rev Mol Cell Biol. Author manuscript; available in PMC 2007 August 17.
You might also like
- Molecular Biology of The Cell, 5th EditionDocument82 pagesMolecular Biology of The Cell, 5th EditionBee Nunes25% (67)
- Active Reading: Answer KeyDocument3 pagesActive Reading: Answer KeyJonas Montero100% (2)
- Bio Calculus PDocument738 pagesBio Calculus PMarco Sousa100% (1)
- Molecular Population Genetics and Evolution - Masatoshi NeiDocument290 pagesMolecular Population Genetics and Evolution - Masatoshi NeiGeorge Sebastian Antony100% (3)
- Extended Heredity: A New Understanding of Inheritance and EvolutionFrom EverandExtended Heredity: A New Understanding of Inheritance and EvolutionNo ratings yet
- PHYS 444 - Physical Biology: From Molecules To Cells Fall 2014 Course InformationDocument6 pagesPHYS 444 - Physical Biology: From Molecules To Cells Fall 2014 Course InformationAnonymous bZTdTpLNo ratings yet
- Coursework B Science 2014 BiologyDocument4 pagesCoursework B Science 2014 Biologyafiwjsazh100% (2)
- Cbe 513Document11 pagesCbe 513duverney.gaviriaNo ratings yet
- Applications of Mathematics To Biology: Manuscript ID: #0443 Original Research PaperDocument5 pagesApplications of Mathematics To Biology: Manuscript ID: #0443 Original Research Paperrizka arzitaNo ratings yet
- The Crossroads Between Biology and Mathematics: The Scientific Method As The Basics of Scientific LiteracyDocument8 pagesThe Crossroads Between Biology and Mathematics: The Scientific Method As The Basics of Scientific LiteracySantiago OrtegonNo ratings yet
- Thesis On Evolutionary BiologyDocument4 pagesThesis On Evolutionary Biologydwfz48q3100% (2)
- A Neural-Functionalist Approach To Learning: Texas A&M University, U.S.ADocument22 pagesA Neural-Functionalist Approach To Learning: Texas A&M University, U.S.AAna HernandezNo ratings yet
- Literature Review Molecular BiologyDocument7 pagesLiterature Review Molecular Biologyafdtsebxc100% (1)
- Misconception About Genetic DriftDocument12 pagesMisconception About Genetic DriftNathalia LaseNo ratings yet
- Admin,+10994 Kloser1Document18 pagesAdmin,+10994 Kloser1Tiago GuimarãesNo ratings yet
- Literary Practices of Biology 1Document6 pagesLiterary Practices of Biology 1api-361092487No ratings yet
- Multiple Representations in Biological Education 2013Document394 pagesMultiple Representations in Biological Education 2013Miguel CalderonNo ratings yet
- Essay 1Document7 pagesEssay 1api-361089929No ratings yet
- Science2004-May-Math in BioDocument5 pagesScience2004-May-Math in BioÁfrica RamosNo ratings yet
- Biophysicsconcep00case PDFDocument360 pagesBiophysicsconcep00case PDFaabc100% (1)
- Biophysics Conce P 00 CaseDocument360 pagesBiophysics Conce P 00 Casemagini3649No ratings yet
- Human Biology Topics For Research PapersDocument7 pagesHuman Biology Topics For Research Papersgz9g97ha100% (1)
- Odom Diffusion Osmosis VeryDocument27 pagesOdom Diffusion Osmosis VerycelestinedordorNo ratings yet
- Leszek Konieczny, Irena Roterman-Konieczna, Paweł Spólnik - Systems Biology - Functional Strategies of Living Organisms-Springer (2023) (Z-Lib - Io)Document269 pagesLeszek Konieczny, Irena Roterman-Konieczna, Paweł Spólnik - Systems Biology - Functional Strategies of Living Organisms-Springer (2023) (Z-Lib - Io)Garbage CanNo ratings yet
- Biology Senior Thesis ExamplesDocument8 pagesBiology Senior Thesis Examplesstephaniebenjaminclarksville100% (1)
- Biology and MathematicsDocument7 pagesBiology and MathematicsMaria AnghelacheNo ratings yet
- Mathematical Manipulative Models: in Defense of "Beanbag Biology"Document11 pagesMathematical Manipulative Models: in Defense of "Beanbag Biology"jungck8715No ratings yet
- Garcia2007b PDFDocument39 pagesGarcia2007b PDFMarciNo ratings yet
- The Basics of Biology - C. StoneDocument294 pagesThe Basics of Biology - C. Stonemonicq77100% (1)
- Bio2010, Report in BriefDocument8 pagesBio2010, Report in BriefNational Academies of Science, Engineering, and MedicineNo ratings yet
- Goriely 2017Document652 pagesGoriely 2017Тимофей НестеровNo ratings yet
- Demographic Methods Across The Tree of Life Roberto Salguero Gomez Full ChapterDocument66 pagesDemographic Methods Across The Tree of Life Roberto Salguero Gomez Full Chapterwilliam.belliveau809100% (21)
- SafariDocument3 pagesSafarinerinajonasNo ratings yet
- Chemistry Lessons For Universities A Review of Constructivist IDEASDocument10 pagesChemistry Lessons For Universities A Review of Constructivist IDEASVictor Manuel Enriquez GNo ratings yet
- Progress in Theoretical Biology: Volume 1From EverandProgress in Theoretical Biology: Volume 1Fred M. SnellNo ratings yet
- Evolution in Changing Environments: Some Theoretical Explorations. (MPB-2)From EverandEvolution in Changing Environments: Some Theoretical Explorations. (MPB-2)No ratings yet
- Molecular Biology Dissertation IdeasDocument4 pagesMolecular Biology Dissertation IdeasBuyCollegePaperOnlineUK100% (1)
- Making Sense of Evolution: The Conceptual Foundations of Evolutionary BiologyFrom EverandMaking Sense of Evolution: The Conceptual Foundations of Evolutionary BiologyRating: 4 out of 5 stars4/5 (3)
- Biology Education Thesis PDFDocument5 pagesBiology Education Thesis PDFPaperHelpWritingCanada100% (1)
- Science ReflectionDocument2 pagesScience Reflectionapi-281124194100% (1)
- Human Genetics by SCDocument626 pagesHuman Genetics by SCtaniyaNo ratings yet
- Uses and Abuses of Mathematics in BiologyDocument4 pagesUses and Abuses of Mathematics in Biologymuhammad sami ullah khanNo ratings yet
- Biologia Matemática 2017Document20 pagesBiologia Matemática 2017iago.sk66No ratings yet
- How To Write A Biology Literature ReviewDocument7 pagesHow To Write A Biology Literature Reviewaiqbzprif100% (1)
- Thesis MicrobiologyDocument8 pagesThesis Microbiologywyppfyhef100% (2)
- The University of Chicago Press Philosophy of Science AssociationDocument11 pagesThe University of Chicago Press Philosophy of Science AssociationAbuAbdur-RazzaqAl-MisriNo ratings yet
- Introductory Physics For The Life Sciences by Simon MochrieDocument860 pagesIntroductory Physics For The Life Sciences by Simon MochrieAina Horrach AlonsoNo ratings yet
- Alberts Introductory PagesDocument33 pagesAlberts Introductory PagesMarianne GonzalesNo ratings yet
- Hugh Drummond (Auth.), Paul Patrick Gordon Bateson, Peter H. Klopfer (Eds.) - Perspectives in Ethology - Volume 4 Advantages of Diversity (1981, Springer US)Document253 pagesHugh Drummond (Auth.), Paul Patrick Gordon Bateson, Peter H. Klopfer (Eds.) - Perspectives in Ethology - Volume 4 Advantages of Diversity (1981, Springer US)Mauro AraújoNo ratings yet
- Biology Literature ReviewDocument4 pagesBiology Literature Reviewjzczyfvkg100% (1)
- Active Voice in ScienceDocument2 pagesActive Voice in SciencelalaNo ratings yet
- Teaching Evolution & The Nature of Science Farber 2003Document9 pagesTeaching Evolution & The Nature of Science Farber 2003Chrystal CarattaNo ratings yet
- Thesis ZoologyDocument7 pagesThesis Zoologytsfublhld100% (2)
- Information and Living Systems BOOKDocument459 pagesInformation and Living Systems BOOKeduardoazevedo_ccciNo ratings yet
- Biology's Next RevolutionDocument1 pageBiology's Next RevolutionbarrospiresNo ratings yet
- Exploring The Limitations of The Scientific MethodDocument4 pagesExploring The Limitations of The Scientific MethodADNo ratings yet
- Introduction To The Philosophy of Science First PartDocument138 pagesIntroduction To The Philosophy of Science First PartChandra Sekhar Sahu100% (1)
- Coek - Info Molecular Biology 2nd EdnDocument1 pageCoek - Info Molecular Biology 2nd EdniqrarNo ratings yet
- Modeling in Computational Biology and Biomedicine:: A Multidisciplinary Endeavor Draft (April 2012)Document31 pagesModeling in Computational Biology and Biomedicine:: A Multidisciplinary Endeavor Draft (April 2012)Dessi DuckNo ratings yet
- Seafood Production - Our World in DataDocument14 pagesSeafood Production - Our World in DataNaresh Prasad SapkotaNo ratings yet
- LGW (G Ptyfg N3'Ljq Ljqlo +:yf Lnld6) 8Sf) T) - F) Q) DFDocument7 pagesLGW (G Ptyfg N3'Ljq Ljqlo +:yf Lnld6) 8Sf) T) - F) Q) DFNaresh Prasad SapkotaNo ratings yet
- My Philosophy - Greg MatDocument5 pagesMy Philosophy - Greg MatNaresh Prasad SapkotaNo ratings yet
- 2019年01月27日GRE考试真题Document22 pages2019年01月27日GRE考试真题Naresh Prasad SapkotaNo ratings yet
- Happy Painting GuideBookDocument3 pagesHappy Painting GuideBookNaresh Prasad SapkotaNo ratings yet
- Bachelor Curriculum Bioinformatics: (In English)Document12 pagesBachelor Curriculum Bioinformatics: (In English)Naresh Prasad SapkotaNo ratings yet
- Week Two Lesson Summaries - CourseraDocument6 pagesWeek Two Lesson Summaries - CourseraNaresh Prasad SapkotaNo ratings yet
- Manjushree Finance - Stock AnalysisDocument9 pagesManjushree Finance - Stock AnalysisNaresh Prasad SapkotaNo ratings yet
- Back To Week 2 LessonsDocument5 pagesBack To Week 2 LessonsNaresh Prasad SapkotaNo ratings yet
- NEWGREPrepByMagoosh 2017Document469 pagesNEWGREPrepByMagoosh 2017Techo Sunny100% (25)
- Dictyostelium Amoebae Are Free Living Cells With A Remarkable Twist: Under The Stress of StarvationDocument4 pagesDictyostelium Amoebae Are Free Living Cells With A Remarkable Twist: Under The Stress of StarvationNaresh Prasad SapkotaNo ratings yet
- L14: 14 Lectures Past and 11 To Go Part I: The Cell As A Well-Stirred Bioreactor'Document3 pagesL14: 14 Lectures Past and 11 To Go Part I: The Cell As A Well-Stirred Bioreactor'Naresh Prasad SapkotaNo ratings yet
- Fine Tuned Model For Perfect Adaptation'Document14 pagesFine Tuned Model For Perfect Adaptation'Naresh Prasad SapkotaNo ratings yet
- Creation of Membrane Associated MindDocument4 pagesCreation of Membrane Associated MindNaresh Prasad SapkotaNo ratings yet
- Engineering Biology To Do Chemistry: BE.010 Spring 2005 Session # 3 NotesDocument2 pagesEngineering Biology To Do Chemistry: BE.010 Spring 2005 Session # 3 NotesNaresh Prasad SapkotaNo ratings yet
- Goals:: Tips To Writing A Successful Proposal For 6.021JDocument2 pagesGoals:: Tips To Writing A Successful Proposal For 6.021JNaresh Prasad SapkotaNo ratings yet
- D Re D: Waiting Times For Chemical ReactionsDocument2 pagesD Re D: Waiting Times For Chemical ReactionsNaresh Prasad SapkotaNo ratings yet
- Summary Lecture 3: Image by MIT OCWDocument6 pagesSummary Lecture 3: Image by MIT OCWNaresh Prasad SapkotaNo ratings yet
- Ps 1Document2 pagesPs 1Naresh Prasad SapkotaNo ratings yet
- Chia-Pet (Chromatin Interaction Analysis by Paired-End Tag Sequencing)Document13 pagesChia-Pet (Chromatin Interaction Analysis by Paired-End Tag Sequencing)Naresh Prasad SapkotaNo ratings yet
- Slides Module01 UsefulGeneticsLecture1F-SlidesDocument13 pagesSlides Module01 UsefulGeneticsLecture1F-SlidesNaresh Prasad SapkotaNo ratings yet
- l16 NotesDocument10 pagesl16 NotesNaresh Prasad SapkotaNo ratings yet
- Systems Biology: X Y X YDocument2 pagesSystems Biology: X Y X YNaresh Prasad SapkotaNo ratings yet
- I Systems Microbiology (13 Lectures) : The Cell As A Well-Stirred Biochemical Reactor'Document20 pagesI Systems Microbiology (13 Lectures) : The Cell As A Well-Stirred Biochemical Reactor'Naresh Prasad SapkotaNo ratings yet
- Final ProjectDocument14 pagesFinal ProjectShivangi VaidNo ratings yet
- LacusCurtius - Strabo's Geography - Book III Chapter 3Document10 pagesLacusCurtius - Strabo's Geography - Book III Chapter 3A Miguel Simão LealNo ratings yet
- Reading and Writing TOS With ExamDocument2 pagesReading and Writing TOS With Examjoy grace jaraNo ratings yet
- MEETING 5 - Describing PeopleDocument6 pagesMEETING 5 - Describing Peoplerizky.ameliaNo ratings yet
- Team EkowDocument4 pagesTeam EkowDavid Ekow NketsiahNo ratings yet
- Banner Value BeamDocument20 pagesBanner Value BeamryanmackintoshNo ratings yet
- OsiSense XU XUK8TAE1MM12 DocumentDocument6 pagesOsiSense XU XUK8TAE1MM12 Documentحمزة صباحNo ratings yet
- Journal of Pharmaceutical Health Services ResearchDocument2 pagesJournal of Pharmaceutical Health Services ResearchTrần Văn ĐệNo ratings yet
- TR 6Document16 pagesTR 6Hritika RajvanshiNo ratings yet
- Erie Scientific LLC: Globally Harmonized Safety Data Sheet (GHSDS)Document7 pagesErie Scientific LLC: Globally Harmonized Safety Data Sheet (GHSDS)Leni MeifitaNo ratings yet
- SilverCrest SHFD 1350 A1 Deep Fryer AppleDocument128 pagesSilverCrest SHFD 1350 A1 Deep Fryer AppleberjanatNo ratings yet
- Duraplate 8200Document4 pagesDuraplate 8200Carlos VillacrizNo ratings yet
- Topographic Map of Flag PondDocument1 pageTopographic Map of Flag PondHistoricalMapsNo ratings yet
- PA4600 & PA6000 User Manual - Rev GDocument53 pagesPA4600 & PA6000 User Manual - Rev GjatzireNo ratings yet
- OSCP Personal Cheatsheet: September 18Th, 2020 o o o oDocument76 pagesOSCP Personal Cheatsheet: September 18Th, 2020 o o o omahesh yadavNo ratings yet
- Organizational Behavior Human Behavior at Work 14th Edition Newstrom Test BankDocument16 pagesOrganizational Behavior Human Behavior at Work 14th Edition Newstrom Test Bankzaneraymondjpanf100% (28)
- Hit and Miss Brickwork: BDA Design Note 02Document2 pagesHit and Miss Brickwork: BDA Design Note 02SamNo ratings yet
- Asphalt Surfacing To Bridge DecksDocument44 pagesAsphalt Surfacing To Bridge DecksNguyễn Văn MinhNo ratings yet
- Simulated Binar Crossover F Continuous Search Space: Comp SystemsDocument34 pagesSimulated Binar Crossover F Continuous Search Space: Comp SystemsSal Ie EmNo ratings yet
- DC Motors Introduction, Equivalent Circuit, TypesDocument22 pagesDC Motors Introduction, Equivalent Circuit, Typesmalik karim dadNo ratings yet
- ConditionalsDocument16 pagesConditionalsAdalid Pulido AriasNo ratings yet
- Statistics Final Pre TestDocument4 pagesStatistics Final Pre TestLorienelNo ratings yet
- It Cleft PDFDocument280 pagesIt Cleft PDFmeannaNo ratings yet
- 593c0be3230b5002a4a21249 Daniel Guillot Ortiz PDFDocument80 pages593c0be3230b5002a4a21249 Daniel Guillot Ortiz PDFElisa MartinezNo ratings yet
- OmniOMS Output GenerationDocument2 pagesOmniOMS Output Generationnitin.aktivNo ratings yet
- Bombardier Data SheetDocument4 pagesBombardier Data SheetLalu Giat AjahNo ratings yet
- Sub-Project Proposal: Municipality of Kabayan Barangay TAWANGANDocument6 pagesSub-Project Proposal: Municipality of Kabayan Barangay TAWANGANpres carap60% (5)
- MTU 10V1600 DS450: Diesel Generator SetDocument4 pagesMTU 10V1600 DS450: Diesel Generator SetJorge Eraldo Albarran PoleoNo ratings yet
- Fin TechDocument32 pagesFin Techkritigupta.may1999No ratings yet