Professional Documents
Culture Documents
In Situ Hybridization PDF
In Situ Hybridization PDF
Uploaded by
Anonymous ThXYCGkLCLOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
In Situ Hybridization PDF
In Situ Hybridization PDF
Uploaded by
Anonymous ThXYCGkLCLCopyright:
Available Formats
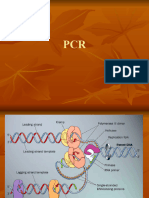
In situ hybridization
In situ hybridization indicates the localization of gene expression in their cellular environment. A labeled RNA or DNA probe
can be used to hybridize to a known target mRNA or DNA sequence within a sample. This labeled RNA or DNA probe can
then be detected by using an antibody to detect the label on the probe. The probes can therefore be used to detect
expression of a gene of interest and the location of the mRNA.
Prepare paraffin sections
RNA/DNA Probe by dehydrating through Frozen
alcohol section
Label by nick
translation
PCR
Labelled RNA/DNA probe Proteinase K treatment of
sections
to expose the DNA/mRNA
Denature Warm slides to
probe hybridization
o
At 95 C temperature
Place 100 l of probe solution onto
tissue section on the slide.
Hybridize the probe to the mRNA or DNA in the
sample
o
55 62 C
Cool to room temperature
Stringency wash to
remove excess
probe
Antibody detection of the probe label
Image
results
Discover more at abcam.com/technical
TIPS:
Storage of samples; RNA preservation in the sample
Preserving DNA is easy because it is a highly stable molecule. However, preserving RNA is much more difficult due to
presence of RNase enzyme. This may be found on glassware, reagents and on the operator and their clothing. RNase will
quickly destroy any RNA in the cell or the RNA probe itself. Therefore users must ensure they use sterile techniques,
gloves, and solutions to prevent RNase from contaminating and destroying the probe or tissue RNA.
General sample storage when using frozen sections
For good results on older slides, the slides should not be stored dry at room temperature. Store either in 100% ethanol at -
o o o
20 C, or place in a plastic box covered in saran wrap at -20 or -80 C. Slides stored in this way can be used for several
years.
Choice of probe
RNA probes
RNA probes should be between 250 to 1500 bases in length. Probes approximately 800 bases long exhibit the highest
sensitivity and specificity. Ideally transcription templates should allow for transcription of both probe (antisense strand) and
negative control (sense strand) RNAs. Cloning into a vector with opposable promoters will achieve this. Circular templates
must be linearized before making a probe. PCR templates can also be used for this purpose.
DNA probes:
DNA probes can also provide high sensitivity to RNA probes. However, they do not hybridize as strongly to the target
mRNA molecules. Therefore, formaldehyde should not be used in the post hybridization washes when using DNA probes.
Specificity of the probe is extremely important. If the exact nucleotide sequence of the mRNA or DNA in the cell is known, a
precise complementary probe can be designed. If over 5 percent of the base pairs are not complementary, the probe will
hybridize only loosely to the target sequence. This means the probe is more likely to be washed away during wash steps
and detection steps and the probe may not be detected, or only some of the sites may be detected and the labeling will not
be an accurate representation.
DIG (Digoxigenin) labeled RNA probe In situ hybridization protocol
The protocol shown here describes the use of DIG labeled single stranded RNA probes to detect expression of the gene of
interest in paraffin embedded section. It is a highly sensitive technique.
1. Deparaffinization
If using formaldehyde fixed paraffin embedded sections.
For frozen sections, please start at section 2
Before proceeding with the staining protocol, the slides must be deparaffinized and rehydrated. Incomplete removal of
paraffin can cause poor staining of the section.
Materials and reagents
Xylene
100% ethanol
95% ethanol
Method
Place the slides in a rack, and perform the following washes:
1. Xylene: 2 x 3 mins.
2. Xylene 1:1 with 100% ethanol: 3 mins.
3. 100% ethanol: 2 x 3 mins.
Discover more at abcam.com/technical
4. 95% ethanol: 3 mins.
5. 70 % ethanol: 3 mins.
6. 50 % ethanol: 3 mins.
7. Running cold tap water to rinse.
Keep the slides in the tap water until ready to perform antigen retrieval. At no time from this point onwards should the slides
be allowed to dry. Drying out will cause non-specific antibody binding and therefore high background staining.
2. Antigen retrieval
o
Digest with 20 g/ml proteinase K in prewarmed 50 mM Tris for 10 to 20 min 37 C. The time of incubation and
concentration of proteinase K may require some optimization.
The concentration of proteinase K and the incubation time for this step will require optimization.
We can recommend trying a proteinase K titration experiment to determine the optimal conditions.
Insufficient digestion will result in a reduced hybridization signal. Over digestion will result in poor
tissue morphology, making localization of the hybridization signal very difficult. The concentration
of Proteinase K needed will vary depending upon the tissue type, length of fixation, and size of tissue.
3. Rinse slides 5 times in distilled water.
4. Immerse slides in ice cold 20% (v/v) acetic acid for 20 seconds. This will permeabilize the cells to allow access to the
probe and the antibody.
5. Dehydrate the sections by washing for approximately 1 min each wash in 70% EtOH, 95% EtOH and 100% EtOH then
air dry.
6. Add 100 l hybridization solution to each section.
Reagent Final conc Amount to use per 1 ml of solution
Formamide 50% 500 l
Salts 5x 250 l
Denhardts solution 5x 100 l
Dextran sulphate 10% 200 l
Heparin 20 U / ml 10 l
SDS 0.1% 10 l
Salt solution:
4 M NaCl
100 mM EDTA
200 mM Tris-HCl pH 7.5
100 mM NaH2PO4.2H2O
100 mM NaH2PO4
Denhardts solution (100x):
10 g Ficoll
10 g PVD (polyvinylpyrrolidine)
10 g BSA (Bovine Serum Albumin)
500ml sterile dH2O
7. Incubate the slides 1 hour in hybridization chamber at the desired hybridization temperature. Typical hybridization
o
temperatures range between 55 and 62 C (for more details, see notes with section 9 below)
o
8. Dilute the probes in hybridization solution ready in PCR tubes. Heat for 95 C for 2 min on a PCR block. This will
dehybridize the RNA or DNA probe. Chill on ice immediately to prevent rehybridization.
Discover more at abcam.com/technical
9. Drain off the hybridization solution. Add 50 to 100 ul per section of diluted probe (ensure the entire sample is covered).
o
Incubate in the hybridization chamber 65 C overnight. Whilst incubating, the sample on the slide can be covered with a
coverslip to prevent evaporation.
During this step, the RNA probe will hybridize to the corresponding mRNA, or the DNA probe will hybridize to the
corresponding cellular DNA.
The hybridization temperature will require optimization depending on the sequence of the probe used, as well
as the cell / tissue type. This temperature should be optimized for each tissue type analyzed.
o
Hybridization temperatures used range from 55 to 62 C.
The optimal hybridization temperature for the probe depends on the percentage of bases present
in the target sequence. The concentration of cytosine and guanine in the sequence are an important factor.
10. Stringency washes:
Solution parameters such as temperature, salt and/or detergent concentration can be manipulated to remove any non-
identical interactions (i.e. only exact sequence matches will remain bound).
To prepare 1 liter of 20 x SSC:
For 1 liter:
175.3 g NaCl (3 M)
88.2 g Na citrate
800 ml sterile dH2O
Adjust to pH 5 using Citric acid, top up to 1 liter and then autoclave.
Wash 1 50% formamide / 2 x SSC
o
3 x for 5 min, 37- 45 C.
o
To wash away any excess probe and the hybridization buffer. Higher temperatures (up to 65 C) can be used for short
periods of time, but this can wash off too much of the hybridiized probe RNA / DNA if left for too long.
Wash 2 0.1-2 x SSC
o o
3 x for 5 min, 25 C to 75 C.
This step removes non-specific and/or repetitive DNA / RNA hybridization. The less concentrated the salt solution and the
longer the duration of the wash and the temperature, the higher the stringency and the more DNA / RNA will be removed.
Optimization of temperatures for stringency washes can be difficult to work out, but the following
guidelines can help:
Very short DNA/RNA probes (0.5-3 kb) or very complex probes, the washing temperature should be lower
O
(up to 45 C) and the stringency lower (1x-2 x SSC).
o
Single-locus or large probes, the temperature should be around 65 C and the stringency high (below 0.5 x SSC).
The temperature and stringency should be highest for repetitive probes (such as alpha-satellite repeats).
11. Wash twice in MABT (maleic acid buffer containing Tween 20) for 30 min at room temperature.
MABT is gentler than PBS and is more suitable for nucleic acid detection.
5 x MABT stock:
500 ml maleic acid pH 7.5
750 mM NaCl
0.5% v/v Tween 20
pH 7.5
12. Dry the slides.
13. Transfer to a humidified chamber and add 200 l blocking buffer to each section (MABT + 2% BSA, milk or serum).
Block for 1 to 2 hours, room temperature.
Discover more at abcam.com/technical
14. Drain off the blocking buffer. Add the anti-label antibody the required dilution in blocking buffer. Check the antibody
datasheet for a recommended concentration. Incubate for 1 to 2 hours at room temperature.
15. Wash slides 5 times with MABT, 10 minutes for each wash, room temperature.
16. Wash the slides 2 x for 10 min room temperature with prestaining buffer (100 mM Tris pH 9.5, 100 mM NaCl, 10 mM
MgCl2).
17. Fluorescence please go next to step 18.
18. Other return slides to humidity chamber and follow manufacturers instructions for color development.
19. Rinse slides in distilled water.
20. Air dry the slides for around 30 minutes. Wash in 100% ethanol, then air dry thoroughly.
21. Mount using DePeX mounting solution.
Discover more at abcam.com/technical
You might also like
- Biology FinalDocument47 pagesBiology FinalAshley Ann StocktonNo ratings yet
- Lab Report On Gene Cloning and Vector ExpressionDocument11 pagesLab Report On Gene Cloning and Vector Expressionselina_kolls100% (2)
- In Situ HybridizationDocument6 pagesIn Situ Hybridizationn7s77hxzbtNo ratings yet
- CTAB ExtractionDocument3 pagesCTAB ExtractionJanikaa Singaravel MuruganNo ratings yet
- PCR Lab ProtocolDocument5 pagesPCR Lab Protocolhk8atema1lNo ratings yet
- Practical Biotech MSCDocument6 pagesPractical Biotech MSCEr Parshant SaharanNo ratings yet
- DNA and RNA ExtractionDocument19 pagesDNA and RNA ExtractionDr. PremalathaNo ratings yet
- 3bsmt1 Bobier, Ashwelldonne Molbio PCRDocument9 pages3bsmt1 Bobier, Ashwelldonne Molbio PCRAshwell Donne BobierNo ratings yet
- Identification of FungiDocument5 pagesIdentification of FungiYến NguyễnNo ratings yet
- PCRHand OutsDocument8 pagesPCRHand OutsMehmood 247No ratings yet
- CH1801 C5 Plasmid IsolationDocument7 pagesCH1801 C5 Plasmid IsolationTanisha ChowdharyNo ratings yet
- Deoxyribonucleic Acid (DNA), Sodium Salt (D1626) - Product Information SheetDocument2 pagesDeoxyribonucleic Acid (DNA), Sodium Salt (D1626) - Product Information SheetJulioNo ratings yet
- Departments: SDMVM'S College of Agricultural BiotechnologyDocument45 pagesDepartments: SDMVM'S College of Agricultural BiotechnologyPAWANKUMAR S. K.No ratings yet
- Southern Northern and Western BlottingDocument40 pagesSouthern Northern and Western BlottingBalaji NaiduNo ratings yet
- DNA Quantification ProtocolDocument4 pagesDNA Quantification Protocolme_dayakar100% (1)
- PCR RTPCR, QPCRDocument88 pagesPCR RTPCR, QPCRshravani sahuNo ratings yet
- Basic Principle: To Study The Isolation of Plant Genomic DNA by Using Modified CTAB MethodDocument35 pagesBasic Principle: To Study The Isolation of Plant Genomic DNA by Using Modified CTAB MethodPAWANKUMAR S. K.No ratings yet
- Preparation of Genomic DNA From BacteriaDocument3 pagesPreparation of Genomic DNA From BacteriaRüveyda AkçinNo ratings yet
- Compiled PracticalsDocument92 pagesCompiled PracticalsAnmol KumarNo ratings yet
- D1S80Document10 pagesD1S80mmarrinnaNo ratings yet
- 2X Taq FroggaMixDocument4 pages2X Taq FroggaMixCésar CuadraNo ratings yet
- Lab Report BET305 - Rahmah Hayati Binti Mohd FauziDocument11 pagesLab Report BET305 - Rahmah Hayati Binti Mohd Fauzirahmah hayatiNo ratings yet
- 626 PCR Amplification Gel VisualizationDocument5 pages626 PCR Amplification Gel VisualizationUgnė KasperavičiūtėNo ratings yet
- Lecture 3 4Document140 pagesLecture 3 4ngocnm.bi12-320No ratings yet
- RT-PCR Two-Steps ProtocolDocument13 pagesRT-PCR Two-Steps ProtocolFrancisco MartinezNo ratings yet
- Nucleic Acid Extraction MethodsDocument19 pagesNucleic Acid Extraction Methodsvg04No ratings yet
- Nucleic Acid Extraction MethodsDocument18 pagesNucleic Acid Extraction MethodssunnyNo ratings yet
- INTRODUCTION Isolation of Plant DNADocument23 pagesINTRODUCTION Isolation of Plant DNAkhushbujain7992No ratings yet
- Experimental Design: ImportanceDocument18 pagesExperimental Design: ImportanceOscar Peña AlvaradoNo ratings yet
- Module 2Document26 pagesModule 2anulorance98No ratings yet
- Standard Operating Procedure (SOP) : 1. General ConsiderationsDocument7 pagesStandard Operating Procedure (SOP) : 1. General ConsiderationsAlemayehu Letebo AlbejoNo ratings yet
- Isolasi DNA WatsonDocument59 pagesIsolasi DNA WatsonHimmatul UlyaNo ratings yet
- Lec 5 DNA Extraction and PCRDocument25 pagesLec 5 DNA Extraction and PCRSaif MohamedNo ratings yet
- Genomic Dna Isolation Protocol From BacteriaDocument4 pagesGenomic Dna Isolation Protocol From BacteriaMd Bulbul AhmedNo ratings yet
- Manual T 1020Document8 pagesManual T 1020حميد حميدNo ratings yet
- Lab Techniques + StorageDocument15 pagesLab Techniques + StorageJoseph Paulo L SilvaNo ratings yet
- Filter-Supported Preparation of X Phage DNA: Analytical Biochemistry 175,196-20Document6 pagesFilter-Supported Preparation of X Phage DNA: Analytical Biochemistry 175,196-20Franco SantinNo ratings yet
- PROTOCOL - IRAP and REMAP For RetrotransposonDocument7 pagesPROTOCOL - IRAP and REMAP For RetrotransposonAngela JimenezNo ratings yet
- Artikel NBDocument9 pagesArtikel NBMagano El-FhiraNo ratings yet
- MAN0012656 Genomic DNA Purification UGDocument4 pagesMAN0012656 Genomic DNA Purification UGranaNo ratings yet
- Nucleic Acid TestingDocument55 pagesNucleic Acid TestingShaiji ShahidNo ratings yet
- Isolation and Characterization of DNADocument75 pagesIsolation and Characterization of DNANathaniel CastasusNo ratings yet
- Molecular Biology - Amity University RajasthanDocument13 pagesMolecular Biology - Amity University Rajasthanabash_u1No ratings yet
- Ez-Pcr Mycoplasma Test Kit: Instructions For UseDocument2 pagesEz-Pcr Mycoplasma Test Kit: Instructions For Usenauval mangantjoNo ratings yet
- Laboratory TechniquesDocument50 pagesLaboratory TechniquesmNo ratings yet
- MEGAclear™ KitDocument12 pagesMEGAclear™ KitXiaojie LiuNo ratings yet
- PCRDocument35 pagesPCRk5zo7jNo ratings yet
- Lab 4.isolation of PlasmidDocument7 pagesLab 4.isolation of PlasmidJane MargarethaNo ratings yet
- Formaldehyde Gel Electrophoresis of Total RNADocument4 pagesFormaldehyde Gel Electrophoresis of Total RNAGhanshyam R ParmarNo ratings yet
- General Biology Laboratory Report: Background: Bacteria Morphology Can Be Divided Into Different Types Based OnDocument7 pagesGeneral Biology Laboratory Report: Background: Bacteria Morphology Can Be Divided Into Different Types Based OnNẻro OwONo ratings yet
- Jove Protocol Multiplex PCR and Reverse Line Blot Hybridization Assay MPCRRLBDocument5 pagesJove Protocol Multiplex PCR and Reverse Line Blot Hybridization Assay MPCRRLBrajeshkundapur123No ratings yet
- Handout For Workshop (DAY 01) (KSBT)Document7 pagesHandout For Workshop (DAY 01) (KSBT)Michael KahnwaldNo ratings yet
- 03 PCRDocument5 pages03 PCRToni D.No ratings yet
- DNA Extraction: Qualitative Estimation of Genomic DNADocument32 pagesDNA Extraction: Qualitative Estimation of Genomic DNAPAWANKUMAR S. K.No ratings yet
- CSI Polymerase Chain Reaction Lab ManualDocument6 pagesCSI Polymerase Chain Reaction Lab ManualDank MoviesNo ratings yet
- TOADERVasile Alin - Paper - 5 - 05 - 2009 - TOADER - 2009Document6 pagesTOADERVasile Alin - Paper - 5 - 05 - 2009 - TOADER - 2009mihaela_neacsuNo ratings yet
- Pzero CloningDocument32 pagesPzero Cloningmaximilianocsc2No ratings yet
- Pic RenderDocument3 pagesPic Renderprateekc29100% (2)
- Standard methods for the examination of water and sewageFrom EverandStandard methods for the examination of water and sewageNo ratings yet
- 6 Molecular Basis of Inheritance - NotesDocument9 pages6 Molecular Basis of Inheritance - NotesAditya Singh0% (1)
- Mutation, DNA Repair, and DNA Integrity - Learn Science at ScitableDocument3 pagesMutation, DNA Repair, and DNA Integrity - Learn Science at ScitableRovin RamphalNo ratings yet
- BS en 12687 - 1998Document12 pagesBS en 12687 - 1998Emanuele MastrangeloNo ratings yet
- Principles of Genetics Maximum Marks: 150 Time: 6 Hrs Section-A Answer All The Questions (20 1 20) Fill in The BlanksDocument5 pagesPrinciples of Genetics Maximum Marks: 150 Time: 6 Hrs Section-A Answer All The Questions (20 1 20) Fill in The BlanksRAJESHNo ratings yet
- Restriction EnzymeDocument45 pagesRestriction Enzymedrsubhash81No ratings yet
- Protein Synthesis: The Protein-Making ProcessDocument21 pagesProtein Synthesis: The Protein-Making ProcessLanie De la Torre100% (1)
- Hyperchromic EffectDocument7 pagesHyperchromic EffectvictorNo ratings yet
- Class 12 Biology Zoology EM Chapter 5. Molecular Genetics Question Bank by Rana Sakthi Durga, Puducherry.Document2 pagesClass 12 Biology Zoology EM Chapter 5. Molecular Genetics Question Bank by Rana Sakthi Durga, Puducherry.joshan jayapaul pandiyan100% (1)
- Molecular MarkersDocument37 pagesMolecular MarkersJeremiah Christopher ManzanoNo ratings yet
- Protein SynthesisDocument6 pagesProtein Synthesishwxmyoief100% (2)
- Jul 2020 MODERNAUS107037891Document248 pagesJul 2020 MODERNAUS107037891Mas ThulanNo ratings yet
- Methylpseudouridylation of mRNA Causes +1 Ribosomal FrameshiftingDocument21 pagesMethylpseudouridylation of mRNA Causes +1 Ribosomal FrameshiftingmeNo ratings yet
- Methods and Applications For Single-Cell and Spatial Multi-OmicsDocument22 pagesMethods and Applications For Single-Cell and Spatial Multi-OmicsXin XuNo ratings yet
- Recognized That Each Individual Has A Unique Pattern of MinisatellitesDocument4 pagesRecognized That Each Individual Has A Unique Pattern of Minisatelliteskeijanne marie mendozaNo ratings yet
- RNA PowerPointDocument20 pagesRNA PowerPointAnki0391100% (1)
- Animal Trans Genesis and CloningDocument230 pagesAnimal Trans Genesis and CloningJanapareddi VenkataramanaNo ratings yet
- Activity Genetic CodeDocument3 pagesActivity Genetic CodeVanessa JuatcoNo ratings yet
- Baltimore ClassificationDocument3 pagesBaltimore ClassificationMarc Allan100% (1)
- Gene SilencingDocument22 pagesGene SilencingDebadrita DasguptaNo ratings yet
- Molecular Basis of Heredity-Ppt3 - SDocument22 pagesMolecular Basis of Heredity-Ppt3 - SJarrah LumpayNo ratings yet
- Short Technical ReportDocument5 pagesShort Technical ReportSato ObcianaNo ratings yet
- Small Molecule Tools To ModulaDocument329 pagesSmall Molecule Tools To Modula吴善统No ratings yet
- Module 4. Nucleic AcidsDocument3 pagesModule 4. Nucleic AcidsKenneth Roy MatuguinaNo ratings yet
- pCDFDuet-1 Map PDFDocument2 pagespCDFDuet-1 Map PDFesn_kNo ratings yet
- TOCE Seegene-ArtigoCientificoDocument16 pagesTOCE Seegene-ArtigoCientificoAna AlonsoNo ratings yet
- Dynamical Transition and Heterogeneous Hydration Dynamics in RNADocument14 pagesDynamical Transition and Heterogeneous Hydration Dynamics in RNAcrocoaliNo ratings yet
- Bioinformatics: Applications NoteDocument2 pagesBioinformatics: Applications NoteRicardo GarcíaNo ratings yet
- Understanding The Basics of DNA Fingerprinting in Forensic ScienceDocument4 pagesUnderstanding The Basics of DNA Fingerprinting in Forensic SciencevivekNo ratings yet