Professional Documents
Culture Documents
Kimball Et Al 2018, Figure 7
Kimball Et Al 2018, Figure 7
Uploaded by
Abby KimballOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Kimball Et Al 2018, Figure 7
Kimball Et Al 2018, Figure 7
Uploaded by
Abby KimballCopyright:
Available Formats
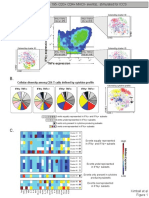
A. viSNE SPADE X-shift Citrus Phenograph B.
% of events
ViSNE/Boolean SPADE
CD45
CD4+
CD8+
CD19+
CD11b+ CD64+
CD11b+ CD64-
X-shift Phenograph CD45+
CD19
CD45-
NKp46+
C. # of clusters/nodes in phenotype
CD4
X-shift Phenograph SPADE
CD4+
CD8+
CD19+
CD11b+ CD64+
CD11b+ CD64-
CD45+
CD45-
NKp46+
CD8
Total=45 Total=29 Total=45
D. 6
Citrus, daughter #82243
SPADE, node #1
X-shift, cluster #515
Phenograph, cluster #8
Median Expression
CD11c
2
NKp46
Tbet
Tim3
Lag3
IkBa
CD19
NKp46
Ki67
PDL2
CD64
PDL1
IRF4
Sig F
FoxP3
KLRG1
Bcatenin
CD127
CD11c
GITR
MHCI
CD69
CD8
CD11b
CD25
Gr-1
p4eBP1
CD3
CTLA4
PD1
Sca1
CD45
CD44
CD4
CD117
MHCII
ICOS
E. 117
MHC
II ICOS Tbet Tim3
Lag
3
IkB
CD a
4 CD
CD 19
4
4
CD
NK
CD64
p4
45
6
CD
Ki6
7 P
1
Sca
DL2
PD1
CD64 PDL1 IRF4 SigF
Citrus, daughter #82243
CTLA4
SPADE, node #1
X-shift, cluster #515
CD11b
Phenograph, cluster #8
CD3
BP1
F
p4e
oxP
3
-1
KL
Gr
viSNE X-shift Phenograph
G1 R
F.
5
D2
Bc
C at
en
b in
D11 CD
C 127
CD8
2,000
CD1
CD69 MHCI GITR 1c
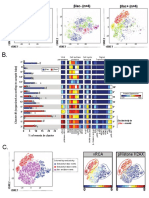
G.
tSNE2
19 clusters tSNE1 20 clusters 50
# of input events per sample
# of Defined Clusters
40 X-shift
5,000
30
tSNE2
23 clusters tSNE1 28 clusters
20
PhenoGraph
10
9,141 #1
0
tSNE2
00
00
#1
#2
00
tSNE1
32 clusters 29 clusters
0
,0
1
1
2,
5,
14
14
20
9,
9,
# of input events per sample
9,141 #2
H.
tSNE2
% of events
32 clusters tSNE1
tSNE130 clusters 0 5 10 15 20 # of
Range clusters <1%
# of input events per sample
2,000 (0.42-15.8) 3 of 20
20,000
Incompatible
5,000 (0.48-15.7) 5 of 28
setting for
algorithm
tSNE2
tSNE1
9,141 #1 (0.14-11.8) 7 of 29
46 clusters 37 clusters
9,141 #2 (0.02-9.83) 9 of 30
(1,263,690)
All events
Insufficient Insufficient 20,000 (0.02-7.96) 14 of 37
processing processing
power power 1%
Figure 7
Kimball et al
You might also like
- Fender Mustang ShematicDocument12 pagesFender Mustang Shematicyebaim100% (4)
- DIAGRAMA Cat c15 BiturboDocument2 pagesDIAGRAMA Cat c15 Biturbohecazorla80% (5)
- Immune Cell Markers PosterDocument1 pageImmune Cell Markers Postercheryyy0% (1)
- Mustang Iv: SMPS + AMP Schematic Diagram 1/3Document12 pagesMustang Iv: SMPS + AMP Schematic Diagram 1/3Valentin NichiforNo ratings yet
- YH305D V6-4 SCH ALLDocument1 pageYH305D V6-4 SCH ALLsibajnk0% (1)
- Psu SchematicsDocument1 pagePsu SchematicsJerry LeeNo ratings yet
- Kimball Et Al 2018, Figure 9Document1 pageKimball Et Al 2018, Figure 9Abby KimballNo ratings yet
- Montarbo 440Document1 pageMontarbo 440AliMudofarNo ratings yet
- 2piling Barge Khang Duc 80 (KD80) DrawingDocument1 page2piling Barge Khang Duc 80 (KD80) DrawingPhú Vũ TrươngNo ratings yet
- Kimball Et Al 2018, Figure 8Document1 pageKimball Et Al 2018, Figure 8Abby KimballNo ratings yet
- +/-0-15V, 0-1A CV/CC Lab Power Supply: PIQ103 PIU201 PIU3012 PIU3012 PIQ103 PIU201Document1 page+/-0-15V, 0-1A CV/CC Lab Power Supply: PIQ103 PIU201 PIU3012 PIU3012 PIQ103 PIU201marco9991No ratings yet
- Canaleta 1Document1 pageCanaleta 1AXEL TUESTA BOCANEGRANo ratings yet
- NCCN Rectal Cancer Guidelines 2018 Benson 874 901Document1 pageNCCN Rectal Cancer Guidelines 2018 Benson 874 901Nikos PapaioannouNo ratings yet
- E01 - Modifikasi sf6Document7 pagesE01 - Modifikasi sf6Carvin AgriantoNo ratings yet
- DD PD DDP PDP QD 10+-550+ Filters Metric Dimension Drawing 9827846800 01Document1 pageDD PD DDP PDP QD 10+-550+ Filters Metric Dimension Drawing 9827846800 01Elías Alejandro Navarro LeónNo ratings yet
- Eon 615Document15 pagesEon 615Uirajara CostaNo ratings yet
- Plano Eléctrico 5500Document36 pagesPlano Eléctrico 5500Johan doradoNo ratings yet
- La Bikina Viola 2Document1 pageLa Bikina Viola 2Jennifer GonzálezNo ratings yet
- Opening Credits PDFDocument2 pagesOpening Credits PDFugurillaNo ratings yet
- Opening Credits: Star Trek:PicardDocument2 pagesOpening Credits: Star Trek:PicardugurillaNo ratings yet
- 1piling Barge Khang Duc 80 (KD80) DrawingDocument1 page1piling Barge Khang Duc 80 (KD80) DrawingPhú Vũ TrươngNo ratings yet
- Great Is Thy Faithfulness - Gibc Orch - 04 - Flute 2Document2 pagesGreat Is Thy Faithfulness - Gibc Orch - 04 - Flute 2Luth ClariñoNo ratings yet
- Pdfslide - Tips - Base Amp Art Series pst355cDocument2 pagesPdfslide - Tips - Base Amp Art Series pst355cvadimNo ratings yet
- Ac Floor Connection SerialDocument1 pageAc Floor Connection Serialانعام مىمونNo ratings yet
- Estructuras-Losa AligeradaDocument1 pageEstructuras-Losa Aligeradahector vera dueñasNo ratings yet
- Music From Gladiator: 1st Clarinet in BDocument2 pagesMusic From Gladiator: 1st Clarinet in BMaestro Valdyr FilhoNo ratings yet
- Carvin Pb100 SCHDocument1 pageCarvin Pb100 SCHАлексей ДеминNo ratings yet
- E360 Schematics Rev20 051201Document9 pagesE360 Schematics Rev20 051201yura19100% (2)
- Manhattanvignettes tb3Document4 pagesManhattanvignettes tb3howardx1220No ratings yet
- YH305D V6-4 SCH ALL PDFDocument1 pageYH305D V6-4 SCH ALL PDFsibajnkNo ratings yet
- Chacarera - Del - Violin - SOLISTA Y QUINTETO-ViolaDocument1 pageChacarera - Del - Violin - SOLISTA Y QUINTETO-ViolaFiorella ScheihingNo ratings yet
- TE NECESITO - Tpta 1Document2 pagesTE NECESITO - Tpta 1GIOVANINo ratings yet
- eDP To LVDSDocument2 pageseDP To LVDSeriy2010sNo ratings yet
- March: Dalla Suite "Lo Schiaccianoci"Document2 pagesMarch: Dalla Suite "Lo Schiaccianoci"Boogie HarryNo ratings yet
- Schematic Half-Bridge ModuleDocument1 pageSchematic Half-Bridge ModuleVitaliyGNo ratings yet
- Caligari Scenes (Cello Part)Document3 pagesCaligari Scenes (Cello Part)topologyrobNo ratings yet
- Mambo 5-Trumpet - in - BB - 1 PDFDocument1 pageMambo 5-Trumpet - in - BB - 1 PDFJohnSéRojasNo ratings yet
- CUATRO PUNTOS CARDINALES - CARNAVALITO-PianoDocument1 pageCUATRO PUNTOS CARDINALES - CARNAVALITO-PianoAlbin.NelsonNo ratings yet
- Great Is Thy Faithfulness - Gibc Orch - 13 - Violin IIDocument3 pagesGreat Is Thy Faithfulness - Gibc Orch - 13 - Violin IILuth ClariñoNo ratings yet
- Trio Telemann Clavecin Sol MineurDocument4 pagesTrio Telemann Clavecin Sol Mineursimonacas94No ratings yet
- A Toda Mecha: (Santa Justa Klan)Document3 pagesA Toda Mecha: (Santa Justa Klan)mmccmmNo ratings yet
- Amplificador Veicular dps2900Document2 pagesAmplificador Veicular dps2900Aparecido EletricaNo ratings yet
- Fra' Martino-Frère Feu: D C B ADocument1 pageFra' Martino-Frère Feu: D C B AfpezNo ratings yet
- Dolphins RicDocument1 pageDolphins RicJacob MounterNo ratings yet
- SuperFantástico TromboneDocument1 pageSuperFantástico Trombonekevin williamNo ratings yet
- DTI Cafe Soluble Dani-A3Document1 pageDTI Cafe Soluble Dani-A3Daniela MatsunoNo ratings yet
- Vestel 17ips10-3 LCD Power Supply Schematic PDFDocument4 pagesVestel 17ips10-3 LCD Power Supply Schematic PDFivan bogdanNo ratings yet
- Piratas Del Caribe RI50 BombardinoDocument3 pagesPiratas Del Caribe RI50 BombardinoYakoButlerGarcíaNo ratings yet
- 25 or 6 To 4: ChicagoDocument2 pages25 or 6 To 4: Chicagoelva kroestzchNo ratings yet
- O Come O Come Emmanuel - Flute-Harp Duet - 06 - Violin IDocument3 pagesO Come O Come Emmanuel - Flute-Harp Duet - 06 - Violin IJoris SANTOSNo ratings yet
- TT50Document11 pagesTT50Tognon MarcoNo ratings yet
- Te-2012 DP: Pip201 Pip202 Pip202 Pip201Document1 pageTe-2012 DP: Pip201 Pip202 Pip202 Pip201Yusuf GüngörNo ratings yet
- K06 16223006 Azzahra Raihana PR3Document3 pagesK06 16223006 Azzahra Raihana PR3Azzahra RaihanaNo ratings yet
- Married Life - Violín 1Document1 pageMarried Life - Violín 1STEWARD OSORIO TABORDANo ratings yet
- California Polytechnic State University San Luis Obispo, CA 93407 WWW - Maps.calpoly - EduDocument1 pageCalifornia Polytechnic State University San Luis Obispo, CA 93407 WWW - Maps.calpoly - EduBob SagNo ratings yet
- Proposed Road at Lebak Sultan KudaratDocument1 pageProposed Road at Lebak Sultan KudaratMarl John PinedaNo ratings yet
- 02 AwakeDocument2 pages02 AwakeMariya RyzhkovaNo ratings yet
- Beethovens Mambo No-Flauta PiccoloDocument2 pagesBeethovens Mambo No-Flauta PiccoloJesús CastañedaNo ratings yet
- Homage Main Board Circuit DaigramDocument1 pageHomage Main Board Circuit DaigramNaseeb Ali GoharNo ratings yet
- Kimball Et Al 2019, Figure 7Document1 pageKimball Et Al 2019, Figure 7Abby KimballNo ratings yet
- Oko Et Al 2019, Figure 8Document1 pageOko Et Al 2019, Figure 8Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 2Document1 pageKimball Et Al 2019, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 3Document1 pageKimball Et Al 2019, Figure 3Abby KimballNo ratings yet
- Neuwelt Et Al, Figure 2Document1 pageNeuwelt Et Al, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 4Document1 pageKimball Et Al 2019, Figure 4Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 5Document1 pageKimball Et Al 2018, Figure 5Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 1Document1 pageKimball Et Al 2019, Figure 1Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 9Document1 pageKimball Et Al 2018, Figure 9Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 8Document1 pageKimball Et Al 2018, Figure 8Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 1Document1 pageKimball Et Al 2018, Figure 1Abby KimballNo ratings yet
- Bullock Et Al 2018, Supplemental Figure 5Document1 pageBullock Et Al 2018, Supplemental Figure 5Abby KimballNo ratings yet
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectDocument1 pageGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballNo ratings yet
- Berger Et Al 2019, Figure 2Document1 pageBerger Et Al 2019, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 2Document1 pageKimball Et Al 2018, Figure 2Abby KimballNo ratings yet
- Bullock Et Al 2018, Figure 6Document1 pageBullock Et Al 2018, Figure 6Abby KimballNo ratings yet
- SC14-07 01-02 Hospital Preventive Maintenance Memo-AttachDocument16 pagesSC14-07 01-02 Hospital Preventive Maintenance Memo-AttachRaydoon SadeqNo ratings yet
- FirePro Technical Prospectus PDFDocument28 pagesFirePro Technical Prospectus PDFRANJITHNo ratings yet
- ZTE Product Introduction and Light PON Solution For BalitowerDocument32 pagesZTE Product Introduction and Light PON Solution For BalitowerOrigoAndoraNo ratings yet
- Ticf KKDDV: Kerala GazetteDocument28 pagesTicf KKDDV: Kerala GazetteVivek KakkothNo ratings yet
- WCP Nietzsche OnNatureGodEthicsDocument4 pagesWCP Nietzsche OnNatureGodEthicsmasohaNo ratings yet
- 06 - Heights and DistancesDocument2 pages06 - Heights and DistancesRekha BhasinNo ratings yet
- Art Appreciation FinalDocument7 pagesArt Appreciation FinalJake Donely C. PaduaNo ratings yet
- Introduction To Ipgcl & PPCLDocument31 pagesIntroduction To Ipgcl & PPCLSahil SethiNo ratings yet
- PP Vs Padillo - PSupt. Pinky Sayson - Acog - February 12, 2018Document3 pagesPP Vs Padillo - PSupt. Pinky Sayson - Acog - February 12, 2018Nelson LaurdenNo ratings yet
- Hazardous Areas - Explosionproof Solenoids: Some HistoryDocument5 pagesHazardous Areas - Explosionproof Solenoids: Some HistoryShahram GhassemiNo ratings yet
- 1.load Test On DC Shut MotorDocument5 pages1.load Test On DC Shut Motorg3v5No ratings yet
- Servicio y Reparación GenieDocument268 pagesServicio y Reparación Geniejonny david martinez perezNo ratings yet
- Modbus Communications Reference Guide: SMD Series DrivesDocument32 pagesModbus Communications Reference Guide: SMD Series DrivesTsietsi SeralaNo ratings yet
- Astm D2563-94Document24 pagesAstm D2563-94Santiago AngelNo ratings yet
- MH370 (MAS370) Malaysia Airlines Flight Tracking and History 11-Mar-2014 (KUL - WMKK-PEK - ZBAA) - FlightAwareDocument2 pagesMH370 (MAS370) Malaysia Airlines Flight Tracking and History 11-Mar-2014 (KUL - WMKK-PEK - ZBAA) - FlightAware陳佩No ratings yet
- Jean AttachmentDocument17 pagesJean AttachmentReena VermaNo ratings yet
- Agribusiness: ExamplesDocument4 pagesAgribusiness: ExamplesAngelo AristizabalNo ratings yet
- 10 Traditional Chilean Desserts - Insanely Good PDFDocument1 page10 Traditional Chilean Desserts - Insanely Good PDFMarilynn SpringerNo ratings yet
- PROPOSALDocument62 pagesPROPOSALJam Dela CruzNo ratings yet
- Dopamine D23 Receptor Antagonism Reduces Activity-Based AnorexiaDocument10 pagesDopamine D23 Receptor Antagonism Reduces Activity-Based AnorexiaJeffery TaylorNo ratings yet
- B-Housing and Living - 1Document2 pagesB-Housing and Living - 1Štěpánka OndrůškováNo ratings yet
- Catalogue - Nitoproof 600PFDocument2 pagesCatalogue - Nitoproof 600PFkenneth0129aaNo ratings yet
- Fashion Through The Decades PowerpointDocument11 pagesFashion Through The Decades PowerpointNigar khanNo ratings yet
- DCEM-21xx BrochureDocument4 pagesDCEM-21xx BrochureApply Softtech100% (1)
- Review of Solar Energy Measurement SystemDocument4 pagesReview of Solar Energy Measurement SystemEditor IJRITCC100% (1)
- Pressure Switch MDR5Document4 pagesPressure Switch MDR5Fidelis NdanoNo ratings yet
- Bullous PemphigoidDocument21 pagesBullous PemphigoidChe Ainil ZainodinNo ratings yet
- 7TH Grade Health Unit 3 Study Guide AnswersDocument8 pages7TH Grade Health Unit 3 Study Guide AnswersBader AlmshariNo ratings yet
- Hand Out MEOW PHYF111Document3 pagesHand Out MEOW PHYF111Shreya GuptaNo ratings yet
- Storage Fading of A Commercial 18650 Cell Comprised With NMC LMO Cathode and Graphite AnodeDocument10 pagesStorage Fading of A Commercial 18650 Cell Comprised With NMC LMO Cathode and Graphite AnodeEnrique MachucaNo ratings yet