Professional Documents
Culture Documents
Kimball Et Al 2018, Figure 8
Kimball Et Al 2018, Figure 8
Uploaded by
Abby KimballCopyright:
Available Formats
You might also like
- Service Manual NovAA 800 enDocument90 pagesService Manual NovAA 800 enAnalista de Servicio50% (4)
- Problem 2.1: Solutions To Chapter 2 Exercise ProblemsDocument58 pagesProblem 2.1: Solutions To Chapter 2 Exercise ProblemsGhazi AlArfaj50% (4)
- Problems and Solutions in Medical Physics DiagnosticDocument157 pagesProblems and Solutions in Medical Physics DiagnosticSubhrojitBagchiNo ratings yet
- Marshall Ma100 Ma50 SCHDocument5 pagesMarshall Ma100 Ma50 SCHMarco ZamoranoNo ratings yet
- Mvt. 2 PDFDocument2 pagesMvt. 2 PDFEmile GousseauNo ratings yet
- Caballo - Viejo Trompeta BDocument1 pageCaballo - Viejo Trompeta BWilliams TutayaNo ratings yet
- Free Mason House - Level 1 West 14Document1 pageFree Mason House - Level 1 West 14Keelver De AsisNo ratings yet
- Tub PVC Sap C-7.5 L 124m: Sub Sistema 02Document1 pageTub PVC Sap C-7.5 L 124m: Sub Sistema 02Lisde EspinozaNo ratings yet
- DIDNT WE - With Eb HornDocument45 pagesDIDNT WE - With Eb Horn6n8nbv5rz7No ratings yet
- Mais Grato A TiDocument1 pageMais Grato A TiVithoria FerbonioNo ratings yet
- Đôi - Giày - Lư I - TRPT & A.SaxDocument2 pagesĐôi - Giày - Lư I - TRPT & A.SaxHoàng ThanhNo ratings yet
- ML December 2023Document2 pagesML December 2023Jayesh KumarNo ratings yet
- Denah Lantai 2-3Document1 pageDenah Lantai 2-3WA ODE SITTI AISYAH ANo ratings yet
- Denah Lantai 1Document1 pageDenah Lantai 1WA ODE SITTI AISYAH ANo ratings yet
- Be - Mechanical Engineering - Semester 7 - 2023 - May - Dloc IV Vibration Controls Rev 2019 C SchemeDocument2 pagesBe - Mechanical Engineering - Semester 7 - 2023 - May - Dloc IV Vibration Controls Rev 2019 C SchemeNuman SiddiquiNo ratings yet
- Entre Tus Manos Anonymous-ÓrganoDocument2 pagesEntre Tus Manos Anonymous-ÓrganoDavid Velazquez MoralesNo ratings yet
- Second Floor: Sec A-A: Typ. Portal Frame DetailDocument1 pageSecond Floor: Sec A-A: Typ. Portal Frame Detailaiman hasanNo ratings yet
- Suite N°3 Bach-VioloncelleDocument6 pagesSuite N°3 Bach-VioloncelleLucas ToubeauNo ratings yet
- Cajas 1-8 ArchivoDocument47 pagesCajas 1-8 ArchivoJorge Aurelio Chavez RoqueNo ratings yet
- Cajas 1-8 Archivo FinalDocument60 pagesCajas 1-8 Archivo FinalJorge Aurelio Chavez RoqueNo ratings yet
- Riop Cuja - Planta1 PDFDocument1 pageRiop Cuja - Planta1 PDFNicolas Giron CuervoNo ratings yet
- Ubicacion BañosDocument1 pageUbicacion BañosPaz Carrasco MellaNo ratings yet
- Gypsy Dance No. 1Document2 pagesGypsy Dance No. 1António CorreiaNo ratings yet
- Daewoo 9000btu Service ManualDocument35 pagesDaewoo 9000btu Service Manualhernandez.josedomingo6804No ratings yet
- GA-01-Rev ADocument1 pageGA-01-Rev AMihailo ČabovićNo ratings yet
- Denah Lantai 4Document1 pageDenah Lantai 4WA ODE SITTI AISYAH ANo ratings yet
- Mix Salay 2025 - GBO KGIDocument37 pagesMix Salay 2025 - GBO KGIfrancodelav12huNo ratings yet
- You'Ve Got A Friend in Me - Toy Story-ViolonceloDocument1 pageYou'Ve Got A Friend in Me - Toy Story-VioloncelotiagoNo ratings yet
- Concerning Hobbits-Violino 2Document1 pageConcerning Hobbits-Violino 2Rodolfo Andrade NunesNo ratings yet
- Kiki S - Delivery - Service #4-VioloncheloDocument2 pagesKiki S - Delivery - Service #4-VioloncheloIvo amber SamaNo ratings yet
- A Song For You TenorDocument2 pagesA Song For You TenorBoatman BillNo ratings yet
- Mission Bay Market ReportDocument4 pagesMission Bay Market ReportDunja GreenNo ratings yet
- Serenata Mozart - VLN IIIDocument1 pageSerenata Mozart - VLN IIIleogiamminola12No ratings yet
- ISA Comp. FinalDocument2 pagesISA Comp. FinalLisbeth OrtizNo ratings yet
- AR Detail 01-07Document1 pageAR Detail 01-07EphremHailuNo ratings yet
- Psu SchematicsDocument1 pagePsu SchematicsJerry LeeNo ratings yet
- River Flows in You - CelloDocument1 pageRiver Flows in You - CelloRocío EspinosaNo ratings yet
- (2018!04!16) Bali DL PRB JustificationDocument7 pages(2018!04!16) Bali DL PRB Justificationangga measNo ratings yet
- Graph Year End 1st FEB 2018Document1 pageGraph Year End 1st FEB 2018hunstreteNo ratings yet
- 3.0 LPG 2010 Schematic Rev C 2010-11-26Document1 page3.0 LPG 2010 Schematic Rev C 2010-11-26David CraigNo ratings yet
- Complex Integration: 2.1 Lne Ntegral N The Omplex PlaneDocument4 pagesComplex Integration: 2.1 Lne Ntegral N The Omplex PlanePanbhi DantNo ratings yet
- 171 Sample ChapterDocument17 pages171 Sample ChapterKalyan SinghNo ratings yet
- Denah Lantai Dasar Denah Lantai Atas: Carport - 0.30Document1 pageDenah Lantai Dasar Denah Lantai Atas: Carport - 0.30hqfq685No ratings yet
- Mission Bay Market ReportDocument4 pagesMission Bay Market ReportDunja GreenNo ratings yet
- Tatlong Bibe MarchDocument1 pageTatlong Bibe MarchDennis Matuceño100% (1)
- Typical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051Document1 pageTypical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051GreenNo ratings yet
- Typical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051Document1 pageTypical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051GreenNo ratings yet
- PLANTA Y PERFIL 5Document1 pagePLANTA Y PERFIL 5AmiNo ratings yet
- PPP Procurement in Ireland An Analysis of Tendering Periods 22Document1 pagePPP Procurement in Ireland An Analysis of Tendering Periods 22ThanveerNo ratings yet
- Ejercicios 1,2 y 3Document1 pageEjercicios 1,2 y 3Hillary Irribari GilNo ratings yet
- Ejercicios 1,2 y 3Document1 pageEjercicios 1,2 y 3Hillary Irribari GilNo ratings yet
- Hs-Se40-Intl Rev BDocument1 pageHs-Se40-Intl Rev BSaurabh GuptaNo ratings yet
- Merangin Hydroelectric Power Plant 350 MW: Power Intake Power Intake Trash Rack Beam Welded Assembly ADocument1 pageMerangin Hydroelectric Power Plant 350 MW: Power Intake Power Intake Trash Rack Beam Welded Assembly ATamboli EnergiNo ratings yet
- Kikis Delivery Service - Moms BroomDocument4 pagesKikis Delivery Service - Moms BroomNatalia Music LiveNo ratings yet
- Vivienda AgapitoDocument1 pageVivienda AgapitoIngenieria YungayNo ratings yet
- Digimon Opening ThemeDocument2 pagesDigimon Opening ThemeLuis IvanNo ratings yet
- Diyos Ay Pag-Ibig 1st Horn in FDocument1 pageDiyos Ay Pag-Ibig 1st Horn in FNonjTreborTendenillaToltolNo ratings yet
- Flute: Arranged by Rory MosherDocument2 pagesFlute: Arranged by Rory MosherClaudio DiasNo ratings yet
- SCALE 1:100 001 SCALE 1:100 001: PavementDocument1 pageSCALE 1:100 001 SCALE 1:100 001: PavementGreenNo ratings yet
- Zaphirus 29 PlatsDocument1 pageZaphirus 29 PlatsDavidNo ratings yet
- Sunset Studios - Shopdrawing 45Document1 pageSunset Studios - Shopdrawing 45Keelver De AsisNo ratings yet
- UNI-750-1 Schematic PostReg - GDocument1 pageUNI-750-1 Schematic PostReg - GJorge Caceres100% (1)
- Kimball Et Al 2019, Figure 7Document1 pageKimball Et Al 2019, Figure 7Abby KimballNo ratings yet
- Oko Et Al 2019, Figure 8Document1 pageOko Et Al 2019, Figure 8Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 2Document1 pageKimball Et Al 2019, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 3Document1 pageKimball Et Al 2019, Figure 3Abby KimballNo ratings yet
- Neuwelt Et Al, Figure 2Document1 pageNeuwelt Et Al, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 4Document1 pageKimball Et Al 2019, Figure 4Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 5Document1 pageKimball Et Al 2018, Figure 5Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 1Document1 pageKimball Et Al 2019, Figure 1Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 9Document1 pageKimball Et Al 2018, Figure 9Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 7Document1 pageKimball Et Al 2018, Figure 7Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 1Document1 pageKimball Et Al 2018, Figure 1Abby KimballNo ratings yet
- Bullock Et Al 2018, Supplemental Figure 5Document1 pageBullock Et Al 2018, Supplemental Figure 5Abby KimballNo ratings yet
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectDocument1 pageGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballNo ratings yet
- Berger Et Al 2019, Figure 2Document1 pageBerger Et Al 2019, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 2Document1 pageKimball Et Al 2018, Figure 2Abby KimballNo ratings yet
- Bullock Et Al 2018, Figure 6Document1 pageBullock Et Al 2018, Figure 6Abby KimballNo ratings yet
- AbarisDocument6 pagesAbarisЛеонид Жмудь100% (1)
- Principles of Environmental Science 8th Edition Cunningham Solutions Manual 1Document36 pagesPrinciples of Environmental Science 8th Edition Cunningham Solutions Manual 1kaylanunezorjsgzqmti100% (31)
- Ac Bridge: EEE 457: Measurement & InstrumentationDocument13 pagesAc Bridge: EEE 457: Measurement & InstrumentationTareqNo ratings yet
- DNH-25W Ex Horn SPK - DSP-25EExmN (T)Document1 pageDNH-25W Ex Horn SPK - DSP-25EExmN (T)Li RongNo ratings yet
- Instrument Loop Check Sheet: DCS/Local Indicator & Visual CheckDocument1 pageInstrument Loop Check Sheet: DCS/Local Indicator & Visual CheckMohd A IshakNo ratings yet
- Electrical Distribution Systems DOC1Document25 pagesElectrical Distribution Systems DOC1Ravi TejaNo ratings yet
- String Handling InstructionsDocument7 pagesString Handling Instructionskarim mohamedNo ratings yet
- MT7621 PDFDocument43 pagesMT7621 PDFCami DragoiNo ratings yet
- Aircraft PropulsionDocument5 pagesAircraft PropulsiondaruvuNo ratings yet
- Quant Checklist 434 by Aashish Arora For Bank Exams 2023Document99 pagesQuant Checklist 434 by Aashish Arora For Bank Exams 2023pallabdey4860No ratings yet
- Native Dynamic SQLDocument2 pagesNative Dynamic SQLRupesh PatraNo ratings yet
- Servlets in JavaDocument26 pagesServlets in JavaPratik GandhiNo ratings yet
- Defects in MaterialDocument15 pagesDefects in MaterialSumit Kumar JhaNo ratings yet
- Earth'S Orbit Path Around The Sun EssayDocument3 pagesEarth'S Orbit Path Around The Sun EssaySebastián VegaNo ratings yet
- Ch5 - CPU SchedulingDocument33 pagesCh5 - CPU SchedulingAditya ThakurNo ratings yet
- Caiib Exam Syllabus 2021Document4 pagesCaiib Exam Syllabus 2021senthil kumar nNo ratings yet
- 4MA0 2F MSC Jan 2019 Edexcel IGCSE MathsDocument15 pages4MA0 2F MSC Jan 2019 Edexcel IGCSE MathsPaca GorriónNo ratings yet
- Media Access ProtocolDocument31 pagesMedia Access ProtocolSupriya DessaiNo ratings yet
- Double Bass GRADE 8Document3 pagesDouble Bass GRADE 8rNo ratings yet
- Simtec Silicone PartsDocument70 pagesSimtec Silicone Partssa_arunkumarNo ratings yet
- Tree Traversals (Inorder, Preorder and Postorder)Document4 pagesTree Traversals (Inorder, Preorder and Postorder)Anand DuraiswamyNo ratings yet
- Chemical Kinetics Reaction Mechanism Frederick LindemannDocument10 pagesChemical Kinetics Reaction Mechanism Frederick LindemannSaman AkramNo ratings yet
- ASIC Interview Questions 1Document5 pagesASIC Interview Questions 1ramNo ratings yet
- Flat Plate Collector IntroDocument4 pagesFlat Plate Collector IntrogcrupaNo ratings yet
- Phillips Science of Dental Materials 4 (Dragged) (Dragged) 6Document1 pagePhillips Science of Dental Materials 4 (Dragged) (Dragged) 6asop06No ratings yet
- HF13Document122 pagesHF13HonoluluNo ratings yet
- Checkweigher RFQ FormDocument2 pagesCheckweigher RFQ Formwasim KhokharNo ratings yet
Kimball Et Al 2018, Figure 8
Kimball Et Al 2018, Figure 8
Uploaded by
Abby KimballOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Kimball Et Al 2018, Figure 8
Kimball Et Al 2018, Figure 8
Uploaded by
Abby KimballCopyright:
Available Formats
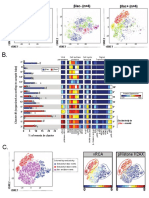
A.
B6 #1 IL10KO #1
B.
40
15.70% 41.80% ***
Colored by:
30
viSNE
CD4+ expression
There are significantly
% CD4+
20 more CD4+ T cells in
IL10KO mice vs. B6 mice.
10
The CD4+ T cell island can be manually 0
gated and % of total events determined. B6 IL10
C. D.
30 B6 Non-significant clusters show
B6 (#1-5) IL10KO (#1-4) high inter-individual variation.
Colored by IL10KO
PhenoGraph
20 cluster ID
% of events
20
0 **
*
-20
tSNE2
-30 0 30-30 0 30
10
tSNE1 Only 2 of 29 PhenoGraph-
defined clusters are
By visual inspection, cluster #4, #5, #8,
identified as significantly
#17, and #25 appear to be different in
different between groups.
cellular abundance between B6 and IL10KO. 0
#4
#5
#8
5
#1
#2
E. B6 biased IL10KO biased
The proportion of events for each F. 100
#504
100
#524
100
#510
cluster from each experimental
% of events in cluster
% of events in cluster
% of events in cluster
100
B6 Biased
80 80 80
B6 condition can be determined.
60 60 60
IL10KO
40 40 40
The biasedness of some clusters
% of events in cluster
20 20 20
X-shift
seem to be driven by individuals. 0 0 0
B6 IL10 B6 IL10 B6 IL10
50
The % of events in each cluster for all #520 #495 #515
IL10KO Biased
individuals can be calculated and 100 100 100
% of events in cluster
% of events in cluster
% of events in cluster
statistical signficance determined.
80 80 80
60 60 60
40 40 40
Of the top 6 clusters most enriched in ** ***
20 20 20
0 either B6 or IL10KO mice, only 2
524
504
510
526
533
503
505
523
511
501
528
502
499
506
509
525
507
498
500
517
527
512
521
497
508
534
496
522
529
532
513
519
531
492
518
514
Input
530
490
493
516
491
494
515
495
520
0 0 0
Cluster ID were statistically significant. B6 IL10 B6 IL10 B6 IL10
G. B6 #1 IL10KO #1 H. * I. B6 #1 IL10KO #1
20 #18 #30 #18 #30
#37
#26 #26 B6 #37
#1 IL10KO #26 #26
#1
ns ns #2 #1 #2 #1
15
% of events
*
SPADE
**
10 *
#9 #12 #9 #12 ***
#31 #31
5
*
** Statistical analysis reveals more nodes
By visual inspection, nodes #1, #9, #12,
0 that are significantly different (#2, #18,
and #26 appear to be different in cellular
#30, #31, and #37) and some that do
1
2
7
8
0
6
#2
#1
#9
abundance between B6 and IL10KO.
#3
#1
#3
#3
#1
#2
Node ID not reach significance (#9, #12).
J. 82268 K. B6 Biased Clusters IL10KO Biased Clusters
#82263 #82265 #82267 #82243 #82252 #82256 #82266
IL10KO biased B6 Biased
82266 82267
1.0 1.0 *** 1.0 ** 1.0 1.0 1.0 1.0
**** **
Abundance
Abundance
Abundance
Abundance
Abundance
Abundance
Abundance
82260 82256 82264 82265 *** ****
0.5 0.5 0.5 0.5 0.5 0.5 0.5
****
82230 82257 82252 82250 82259 82263 82251
0.0 0.0 0.0 0.0 0.0 0.0 0.0
6
IL 6
O
6
O
B
B
B
B
K
K
K
82239 82254 82243 82236 82249 82255 82262 82261 82241
10
10
10
10
10
10
10
IL
IL
IL
IL
IL
IL
Citrus
82234 82232 82228 82247 82258
Citrus is the only algorithm which automatically identifies statistically significant differences.
82223 82229 82253 82238
Abundance values can be extracted from Citrus files to calculate an actual p value.
Cellular phenotype for each cluster can be defined by histogram overlays.
L. Tim3
CD4 CTLA4 GITR ICOS IRF4 Ki67 Lag3 PD1 Tbet
#82243
Background Expression Cluster Expression Figure 8
Kimball et al
You might also like
- Service Manual NovAA 800 enDocument90 pagesService Manual NovAA 800 enAnalista de Servicio50% (4)
- Problem 2.1: Solutions To Chapter 2 Exercise ProblemsDocument58 pagesProblem 2.1: Solutions To Chapter 2 Exercise ProblemsGhazi AlArfaj50% (4)
- Problems and Solutions in Medical Physics DiagnosticDocument157 pagesProblems and Solutions in Medical Physics DiagnosticSubhrojitBagchiNo ratings yet
- Marshall Ma100 Ma50 SCHDocument5 pagesMarshall Ma100 Ma50 SCHMarco ZamoranoNo ratings yet
- Mvt. 2 PDFDocument2 pagesMvt. 2 PDFEmile GousseauNo ratings yet
- Caballo - Viejo Trompeta BDocument1 pageCaballo - Viejo Trompeta BWilliams TutayaNo ratings yet
- Free Mason House - Level 1 West 14Document1 pageFree Mason House - Level 1 West 14Keelver De AsisNo ratings yet
- Tub PVC Sap C-7.5 L 124m: Sub Sistema 02Document1 pageTub PVC Sap C-7.5 L 124m: Sub Sistema 02Lisde EspinozaNo ratings yet
- DIDNT WE - With Eb HornDocument45 pagesDIDNT WE - With Eb Horn6n8nbv5rz7No ratings yet
- Mais Grato A TiDocument1 pageMais Grato A TiVithoria FerbonioNo ratings yet
- Đôi - Giày - Lư I - TRPT & A.SaxDocument2 pagesĐôi - Giày - Lư I - TRPT & A.SaxHoàng ThanhNo ratings yet
- ML December 2023Document2 pagesML December 2023Jayesh KumarNo ratings yet
- Denah Lantai 2-3Document1 pageDenah Lantai 2-3WA ODE SITTI AISYAH ANo ratings yet
- Denah Lantai 1Document1 pageDenah Lantai 1WA ODE SITTI AISYAH ANo ratings yet
- Be - Mechanical Engineering - Semester 7 - 2023 - May - Dloc IV Vibration Controls Rev 2019 C SchemeDocument2 pagesBe - Mechanical Engineering - Semester 7 - 2023 - May - Dloc IV Vibration Controls Rev 2019 C SchemeNuman SiddiquiNo ratings yet
- Entre Tus Manos Anonymous-ÓrganoDocument2 pagesEntre Tus Manos Anonymous-ÓrganoDavid Velazquez MoralesNo ratings yet
- Second Floor: Sec A-A: Typ. Portal Frame DetailDocument1 pageSecond Floor: Sec A-A: Typ. Portal Frame Detailaiman hasanNo ratings yet
- Suite N°3 Bach-VioloncelleDocument6 pagesSuite N°3 Bach-VioloncelleLucas ToubeauNo ratings yet
- Cajas 1-8 ArchivoDocument47 pagesCajas 1-8 ArchivoJorge Aurelio Chavez RoqueNo ratings yet
- Cajas 1-8 Archivo FinalDocument60 pagesCajas 1-8 Archivo FinalJorge Aurelio Chavez RoqueNo ratings yet
- Riop Cuja - Planta1 PDFDocument1 pageRiop Cuja - Planta1 PDFNicolas Giron CuervoNo ratings yet
- Ubicacion BañosDocument1 pageUbicacion BañosPaz Carrasco MellaNo ratings yet
- Gypsy Dance No. 1Document2 pagesGypsy Dance No. 1António CorreiaNo ratings yet
- Daewoo 9000btu Service ManualDocument35 pagesDaewoo 9000btu Service Manualhernandez.josedomingo6804No ratings yet
- GA-01-Rev ADocument1 pageGA-01-Rev AMihailo ČabovićNo ratings yet
- Denah Lantai 4Document1 pageDenah Lantai 4WA ODE SITTI AISYAH ANo ratings yet
- Mix Salay 2025 - GBO KGIDocument37 pagesMix Salay 2025 - GBO KGIfrancodelav12huNo ratings yet
- You'Ve Got A Friend in Me - Toy Story-ViolonceloDocument1 pageYou'Ve Got A Friend in Me - Toy Story-VioloncelotiagoNo ratings yet
- Concerning Hobbits-Violino 2Document1 pageConcerning Hobbits-Violino 2Rodolfo Andrade NunesNo ratings yet
- Kiki S - Delivery - Service #4-VioloncheloDocument2 pagesKiki S - Delivery - Service #4-VioloncheloIvo amber SamaNo ratings yet
- A Song For You TenorDocument2 pagesA Song For You TenorBoatman BillNo ratings yet
- Mission Bay Market ReportDocument4 pagesMission Bay Market ReportDunja GreenNo ratings yet
- Serenata Mozart - VLN IIIDocument1 pageSerenata Mozart - VLN IIIleogiamminola12No ratings yet
- ISA Comp. FinalDocument2 pagesISA Comp. FinalLisbeth OrtizNo ratings yet
- AR Detail 01-07Document1 pageAR Detail 01-07EphremHailuNo ratings yet
- Psu SchematicsDocument1 pagePsu SchematicsJerry LeeNo ratings yet
- River Flows in You - CelloDocument1 pageRiver Flows in You - CelloRocío EspinosaNo ratings yet
- (2018!04!16) Bali DL PRB JustificationDocument7 pages(2018!04!16) Bali DL PRB Justificationangga measNo ratings yet
- Graph Year End 1st FEB 2018Document1 pageGraph Year End 1st FEB 2018hunstreteNo ratings yet
- 3.0 LPG 2010 Schematic Rev C 2010-11-26Document1 page3.0 LPG 2010 Schematic Rev C 2010-11-26David CraigNo ratings yet
- Complex Integration: 2.1 Lne Ntegral N The Omplex PlaneDocument4 pagesComplex Integration: 2.1 Lne Ntegral N The Omplex PlanePanbhi DantNo ratings yet
- 171 Sample ChapterDocument17 pages171 Sample ChapterKalyan SinghNo ratings yet
- Denah Lantai Dasar Denah Lantai Atas: Carport - 0.30Document1 pageDenah Lantai Dasar Denah Lantai Atas: Carport - 0.30hqfq685No ratings yet
- Mission Bay Market ReportDocument4 pagesMission Bay Market ReportDunja GreenNo ratings yet
- Tatlong Bibe MarchDocument1 pageTatlong Bibe MarchDennis Matuceño100% (1)
- Typical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051Document1 pageTypical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051GreenNo ratings yet
- Typical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051Document1 pageTypical Elevtaion 3 Typical Section 4: SCALE 1:50 051 SCALE 1:50 051GreenNo ratings yet
- PLANTA Y PERFIL 5Document1 pagePLANTA Y PERFIL 5AmiNo ratings yet
- PPP Procurement in Ireland An Analysis of Tendering Periods 22Document1 pagePPP Procurement in Ireland An Analysis of Tendering Periods 22ThanveerNo ratings yet
- Ejercicios 1,2 y 3Document1 pageEjercicios 1,2 y 3Hillary Irribari GilNo ratings yet
- Ejercicios 1,2 y 3Document1 pageEjercicios 1,2 y 3Hillary Irribari GilNo ratings yet
- Hs-Se40-Intl Rev BDocument1 pageHs-Se40-Intl Rev BSaurabh GuptaNo ratings yet
- Merangin Hydroelectric Power Plant 350 MW: Power Intake Power Intake Trash Rack Beam Welded Assembly ADocument1 pageMerangin Hydroelectric Power Plant 350 MW: Power Intake Power Intake Trash Rack Beam Welded Assembly ATamboli EnergiNo ratings yet
- Kikis Delivery Service - Moms BroomDocument4 pagesKikis Delivery Service - Moms BroomNatalia Music LiveNo ratings yet
- Vivienda AgapitoDocument1 pageVivienda AgapitoIngenieria YungayNo ratings yet
- Digimon Opening ThemeDocument2 pagesDigimon Opening ThemeLuis IvanNo ratings yet
- Diyos Ay Pag-Ibig 1st Horn in FDocument1 pageDiyos Ay Pag-Ibig 1st Horn in FNonjTreborTendenillaToltolNo ratings yet
- Flute: Arranged by Rory MosherDocument2 pagesFlute: Arranged by Rory MosherClaudio DiasNo ratings yet
- SCALE 1:100 001 SCALE 1:100 001: PavementDocument1 pageSCALE 1:100 001 SCALE 1:100 001: PavementGreenNo ratings yet
- Zaphirus 29 PlatsDocument1 pageZaphirus 29 PlatsDavidNo ratings yet
- Sunset Studios - Shopdrawing 45Document1 pageSunset Studios - Shopdrawing 45Keelver De AsisNo ratings yet
- UNI-750-1 Schematic PostReg - GDocument1 pageUNI-750-1 Schematic PostReg - GJorge Caceres100% (1)
- Kimball Et Al 2019, Figure 7Document1 pageKimball Et Al 2019, Figure 7Abby KimballNo ratings yet
- Oko Et Al 2019, Figure 8Document1 pageOko Et Al 2019, Figure 8Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 2Document1 pageKimball Et Al 2019, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 3Document1 pageKimball Et Al 2019, Figure 3Abby KimballNo ratings yet
- Neuwelt Et Al, Figure 2Document1 pageNeuwelt Et Al, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 4Document1 pageKimball Et Al 2019, Figure 4Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 5Document1 pageKimball Et Al 2018, Figure 5Abby KimballNo ratings yet
- Kimball Et Al 2019, Figure 1Document1 pageKimball Et Al 2019, Figure 1Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 9Document1 pageKimball Et Al 2018, Figure 9Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 7Document1 pageKimball Et Al 2018, Figure 7Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 1Document1 pageKimball Et Al 2018, Figure 1Abby KimballNo ratings yet
- Bullock Et Al 2018, Supplemental Figure 5Document1 pageBullock Et Al 2018, Supplemental Figure 5Abby KimballNo ratings yet
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectDocument1 pageGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballNo ratings yet
- Berger Et Al 2019, Figure 2Document1 pageBerger Et Al 2019, Figure 2Abby KimballNo ratings yet
- Kimball Et Al 2018, Figure 2Document1 pageKimball Et Al 2018, Figure 2Abby KimballNo ratings yet
- Bullock Et Al 2018, Figure 6Document1 pageBullock Et Al 2018, Figure 6Abby KimballNo ratings yet
- AbarisDocument6 pagesAbarisЛеонид Жмудь100% (1)
- Principles of Environmental Science 8th Edition Cunningham Solutions Manual 1Document36 pagesPrinciples of Environmental Science 8th Edition Cunningham Solutions Manual 1kaylanunezorjsgzqmti100% (31)
- Ac Bridge: EEE 457: Measurement & InstrumentationDocument13 pagesAc Bridge: EEE 457: Measurement & InstrumentationTareqNo ratings yet
- DNH-25W Ex Horn SPK - DSP-25EExmN (T)Document1 pageDNH-25W Ex Horn SPK - DSP-25EExmN (T)Li RongNo ratings yet
- Instrument Loop Check Sheet: DCS/Local Indicator & Visual CheckDocument1 pageInstrument Loop Check Sheet: DCS/Local Indicator & Visual CheckMohd A IshakNo ratings yet
- Electrical Distribution Systems DOC1Document25 pagesElectrical Distribution Systems DOC1Ravi TejaNo ratings yet
- String Handling InstructionsDocument7 pagesString Handling Instructionskarim mohamedNo ratings yet
- MT7621 PDFDocument43 pagesMT7621 PDFCami DragoiNo ratings yet
- Aircraft PropulsionDocument5 pagesAircraft PropulsiondaruvuNo ratings yet
- Quant Checklist 434 by Aashish Arora For Bank Exams 2023Document99 pagesQuant Checklist 434 by Aashish Arora For Bank Exams 2023pallabdey4860No ratings yet
- Native Dynamic SQLDocument2 pagesNative Dynamic SQLRupesh PatraNo ratings yet
- Servlets in JavaDocument26 pagesServlets in JavaPratik GandhiNo ratings yet
- Defects in MaterialDocument15 pagesDefects in MaterialSumit Kumar JhaNo ratings yet
- Earth'S Orbit Path Around The Sun EssayDocument3 pagesEarth'S Orbit Path Around The Sun EssaySebastián VegaNo ratings yet
- Ch5 - CPU SchedulingDocument33 pagesCh5 - CPU SchedulingAditya ThakurNo ratings yet
- Caiib Exam Syllabus 2021Document4 pagesCaiib Exam Syllabus 2021senthil kumar nNo ratings yet
- 4MA0 2F MSC Jan 2019 Edexcel IGCSE MathsDocument15 pages4MA0 2F MSC Jan 2019 Edexcel IGCSE MathsPaca GorriónNo ratings yet
- Media Access ProtocolDocument31 pagesMedia Access ProtocolSupriya DessaiNo ratings yet
- Double Bass GRADE 8Document3 pagesDouble Bass GRADE 8rNo ratings yet
- Simtec Silicone PartsDocument70 pagesSimtec Silicone Partssa_arunkumarNo ratings yet
- Tree Traversals (Inorder, Preorder and Postorder)Document4 pagesTree Traversals (Inorder, Preorder and Postorder)Anand DuraiswamyNo ratings yet
- Chemical Kinetics Reaction Mechanism Frederick LindemannDocument10 pagesChemical Kinetics Reaction Mechanism Frederick LindemannSaman AkramNo ratings yet
- ASIC Interview Questions 1Document5 pagesASIC Interview Questions 1ramNo ratings yet
- Flat Plate Collector IntroDocument4 pagesFlat Plate Collector IntrogcrupaNo ratings yet
- Phillips Science of Dental Materials 4 (Dragged) (Dragged) 6Document1 pagePhillips Science of Dental Materials 4 (Dragged) (Dragged) 6asop06No ratings yet
- HF13Document122 pagesHF13HonoluluNo ratings yet
- Checkweigher RFQ FormDocument2 pagesCheckweigher RFQ Formwasim KhokharNo ratings yet