Professional Documents
Culture Documents
######## #2 Preparar Datos
######## #2 Preparar Datos
Uploaded by
LEO KIMIOriginal Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
######## #2 Preparar Datos
######## #2 Preparar Datos
Uploaded by
LEO KIMICopyright:
Available Formats
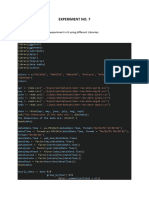
library(caret)
library(pROC)
library(rpart.plot)
library("doParallel")
no_cores <- detectCores()-1
registerDoParallel(cores=no_cores)
cl<-makeCluster(no_cores)
data <- read.csv(file="c:/R/face.csv", header=T, sep=";")
######## #2 PREPARAR DATOS
data$likebinary <- factor(ifelse(data$like>median(data$like), "si", "no"))
data$Type <- factor(data$Type,level=c("Link","Photo","Status","Video"))
data$Paid <- factor(data$Paid,level=c(0,1))
data$Category<- factor(data$Category,level=c(1,2,3))
data$Post.Month<- factor(data$Post.Month,level=c(1:12))
data$Post.Weekday<- factor(data$Post.Weekday,level=c(1:7))
data$Post.Hour<- factor(data$Post.Hour)
attach(data)
df <- data.frame(Type, Category, Post.Month, Post.Weekday, Post.Hour,Paid,likebinary)
######### #1 PARTICIÓN
set.seed(1234)
partition <- createDataPartition(df$likebinary, p=0.7, list=F, times=1)
train <- df[partition,]
test <- df[-partition,]
######### MODELOS
control <-
trainControl(method="cv",number=10,summaryFunction=twoClassSummary,classProbs=T,savePredictions=T)
##optim NB
grid1 <- expand.grid(usekernel = c(T, F), fL = 0:15, adjust = seq(0, 15, by = 1))
set.seed(1234)
m1b <- train(x=subset(train, select=-likebinary),y=train$likebinary, method="nb",trControl=control,tuneGrid = grid1)
plot(m1b)
predicted1b <- predict(m1b, test)
predictedprobs1b <- predict(m1b, test,type="prob")
##optim CT
grid2 <- expand.grid(cp=c(seq(0,.1,.001), seq(.0,1,.1)))
set.seed(1234)
m2b <- train(x=subset(train, select=-likebinary),y=train$likebinary, method="rpart",trControl=control,tuneGrid =
grid2)
plot(m2b)
m2b$finalModel
rpart.plot(m2b$finalModel)
predicted2b <- predict(m2b, test)
predictedprobs2b <- predict(m2b, test,type="prob")
##optim RF
grid3 <- expand.grid(mtry=(1:20))
set.seed(1234)
m3b <- train(x=subset(train, select=-likebinary),y=train$likebinary, method="rf",trControl=control,tuneGrid = grid3)
plot(m3b)
predicted3b <- predict(m3b, test)
predictedprobs3b <- predict(m3b, test,type="prob")
######### #3 COMPARAR MODELOS
cf1b <- confusionMatrix(m1b)
cf2b <- confusionMatrix(m2b)
cf3b <- confusionMatrix(m3b)
cm1b <- confusionMatrix(predicted1b, test$likebinary)
cm2b <- confusionMatrix(predicted2b, test$likebinary)
cm3b <- confusionMatrix(predicted3b, test$likebinary)
rocg1b <- roc(m1b$pred$obs, m1b$pred$si,levels=rev(levels(test$likebinary))))
rocg2b <- roc(m2b$pred$obs, m2b$pred$si,levels=rev(levels(test$likebinary))))
rocg3b <- roc(m3b$pred$obs, m3b$pred$si,levels=rev(levels(test$likebinary))))
plot(rocg3b)
plot(rocg1b)
lines(rocg2b ,col="blue")
lines(rocg3b ,col="red")
legend("bottomright",legend=c("Naive Bayes","Rpart","Bosque aleatorio"),lty=1,col=c("black", "blue","red"),lwd=2)
compareb <- data.frame(
" " = c("Training accuracy","Test accuracy","Test Sensitivity","Test Specificity", "ROC AUC"),
"
"=c(cf1b$table[1,1]/rowSums(cf1b$table)[[1]],cm1b$overall[[1]],cm1b$byClass[[1]],cm1b$byClass[[2]],rocg1b$auc),
"
"=c(cf2b$table[1,1]/rowSums(cf2b$table)[[1]],cm2b$overall[[1]],cm2b$byClass[[1]],cm2b$byClass[[2]],rocg2b$auc),
"
"=c(cf3b$table[1,1]/rowSums(cf3b$table)[[1]],cm3b$overall[[1]],cm3b$byClass[[1]],cm3b$byClass[[2]],rocg3b$auc)
);colnames(compareb) <- c("","Naive Bayes","Classification Tree","Random Forest");compareb
You might also like
- Tarea de Ciencia de DatosDocument32 pagesTarea de Ciencia de DatosLeomaris FerrerasNo ratings yet
- CodesDocument14 pagesCodesArvind NANDAN SINGHNo ratings yet
- Snap GeneDocument728 pagesSnap Genehermann100% (2)
- Association R: AppendixDocument5 pagesAssociation R: AppendixElNo ratings yet
- #Initialising and Loading The LibrariesDocument7 pages#Initialising and Loading The LibrariesGoutam BiswasNo ratings yet
- My Research CodeDocument1 pageMy Research Codewhyyou11184No ratings yet
- Return Calculations in R Return Calculations in R: Econ 424 Fall 2010 Eric Zivot Eric Zivot Updated: October 8, 2010Document13 pagesReturn Calculations in R Return Calculations in R: Econ 424 Fall 2010 Eric Zivot Eric Zivot Updated: October 8, 2010Jaishree ChauhanNo ratings yet
- AIML Lab - Ws10Document9 pagesAIML Lab - Ws10lucky oneNo ratings yet
- DatasetsDocument40 pagesDatasetsAsmatullah KhanNo ratings yet
- Lab 1: Introduction To R: 1 Installing SoftwareDocument4 pagesLab 1: Introduction To R: 1 Installing SoftwareGustavo SánchezNo ratings yet
- GetwdDocument24 pagesGetwdElysa MusarofahNo ratings yet
- Rstudiogg 21Document3 pagesRstudiogg 21Assia GuetniNo ratings yet
- Rstudio MDRDocument4 pagesRstudio MDRAssia GuetniNo ratings yet
- R CommandDocument52 pagesR Commandkoustav nahaNo ratings yet
- PracticalDocument22 pagesPractical786chetanNo ratings yet
- CodeDocument3 pagesCodeYoyo NaNo ratings yet
- RSCH8079 - Session 09 - Data Science With RDocument69 pagesRSCH8079 - Session 09 - Data Science With RDinne RatjNo ratings yet
- 36C3 SQLite3 OmerGullDocument104 pages36C3 SQLite3 OmerGullReid DossingerNo ratings yet
- Python Crash Course by Ehmatthes 16Document1 pagePython Crash Course by Ehmatthes 16alfonsofdezNo ratings yet
- Chapter 2Document16 pagesChapter 2Deepak GahlotNo ratings yet
- Escript Com Rede de CorrelaçãoDocument2 pagesEscript Com Rede de CorrelaçãoRychaellen Silva de BritoNo ratings yet
- LibraryDocument1 pageLibraryEben-ezer AyanouNo ratings yet
- R Syntax Examples 1Document6 pagesR Syntax Examples 1Pedro CruzNo ratings yet
- RG Inference CodeDocument3 pagesRG Inference CodeBrokin HartNo ratings yet
- R Lab ProgramsDocument19 pagesR Lab Programsmdqhizar1211125No ratings yet
- Data Wrangling (Data Preprocessing)Document4 pagesData Wrangling (Data Preprocessing)Siddharth RaulNo ratings yet
- File - Choose - Opens Up The Directory To Select Desired File Read - SCV - Reads The .CSV Format FileDocument3 pagesFile - Choose - Opens Up The Directory To Select Desired File Read - SCV - Reads The .CSV Format Fileabhishekk594No ratings yet
- R语言基础入门指令 (tips)Document14 pagesR语言基础入门指令 (tips)s2000152No ratings yet
- Introduction To RDocument36 pagesIntroduction To RRefael LavNo ratings yet
- SPARKDocument36 pagesSPARKchenna kesavaNo ratings yet
- 10-Visualization of Streaming Data and Class R Code-10!03!2023Document19 pages10-Visualization of Streaming Data and Class R Code-10!03!2023G Krishna VamsiNo ratings yet
- EM622 Data Analysis and Visualization Techniques For Decision-MakingDocument47 pagesEM622 Data Analysis and Visualization Techniques For Decision-MakingRidhi BNo ratings yet
- Likert ProjectDocument2 pagesLikert Projectlawyer thapaNo ratings yet
- CARLDocument3 pagesCARLAli KamranNo ratings yet
- Digital Assignment-6: Read The DataDocument30 pagesDigital Assignment-6: Read The DataPavan KarthikeyaNo ratings yet
- Aman DA 111Document14 pagesAman DA 111Adarsh yadavNo ratings yet
- 03 Data Input OutputDocument43 pages03 Data Input OutputAlexandra Gabriela GrecuNo ratings yet
- R Code ECON ProjDocument10 pagesR Code ECON ProjNora SoualhiNo ratings yet
- JavaDocument7 pagesJavaAzaroathNo ratings yet
- 8Document3 pages8dowemo9017No ratings yet
- Ads Final Assignment No 11Document18 pagesAds Final Assignment No 11ertNo ratings yet
- Codigos RDocument12 pagesCodigos RCarlos Lizárraga RoblesNo ratings yet
- Code Documentation: Loading The PackagesDocument5 pagesCode Documentation: Loading The PackagesShashwat Patel mm19b053No ratings yet
- PSPPTDocument10 pagesPSPPTSowmyaNo ratings yet
- Ps NewDocument9 pagesPs NewSowmyaNo ratings yet
- R Training Deck - v1Document35 pagesR Training Deck - v1Mrigendranath DebsarmaNo ratings yet
- Sintaks Skrip-2Document6 pagesSintaks Skrip-2Hasma Sulaiman SulaimanNo ratings yet
- Metafor TutorialDocument9 pagesMetafor TutorialSurya DilaNo ratings yet
- DAV Practical 7Document3 pagesDAV Practical 7potake7704No ratings yet
- Introduction To Pandas LibraryDocument4 pagesIntroduction To Pandas LibraryAbhisingNo ratings yet
- Exam Cheatsheet For R Langauge CodingDocument2 pagesExam Cheatsheet For R Langauge CodingamirNo ratings yet
- Neural Net WorkDocument2 pagesNeural Net Workspace manNo ratings yet
- Regression With RDocument3 pagesRegression With RfrisbeeeNo ratings yet
- Twittermining: 1 Twitter Text Mining - Required LibrariesDocument4 pagesTwittermining: 1 Twitter Text Mining - Required LibrariesSamuel PeoplesNo ratings yet
- Discrete Math Ass1 CodeDocument2 pagesDiscrete Math Ass1 Codephuongnha922017No ratings yet
- Machine Learning Using R An Introduction To R Mrs. Lijetha C Jaffrin Ap/It VeltechDocument32 pagesMachine Learning Using R An Introduction To R Mrs. Lijetha C Jaffrin Ap/It VeltechlijethaNo ratings yet
- RSTUDIODocument44 pagesRSTUDIOsamarth agarwalNo ratings yet
- J QueryDocument7 pagesJ QuerySaniya PatilNo ratings yet
- DocseDocument3 pagesDocselodakel125No ratings yet
- DAFZA Security - User ManualDocument28 pagesDAFZA Security - User Manualzubeen RIZVINo ratings yet
- TERRAGEN DOFTutorialDocument14 pagesTERRAGEN DOFTutorialCresencio TurpoNo ratings yet
- LATENCYDocument3 pagesLATENCYkatya ZekoviaNo ratings yet
- Introduction To MPLS Technologies: Santanu DasguptaDocument75 pagesIntroduction To MPLS Technologies: Santanu DasguptaĐỗ Tuấn HàoNo ratings yet
- Root & Unroot I9082 v4.2.2Document11 pagesRoot & Unroot I9082 v4.2.2duytrung2121No ratings yet
- Evolution of MediaDocument11 pagesEvolution of MediaJULIUS E DIAZONNo ratings yet
- Optical Paper 3Document9 pagesOptical Paper 3Pir Meher Ali Eng DepartmentNo ratings yet
- InformationSystemAUDIT en-USDocument121 pagesInformationSystemAUDIT en-USKarthickNo ratings yet
- Yale Smart Safe Datasheet EnglishDocument2 pagesYale Smart Safe Datasheet EnglishMarko ĆavarNo ratings yet
- AD831Document16 pagesAD831Vicent Ortiz GonzalezNo ratings yet
- Automobile Tracking System Using Gps and GSM: Jaya Ram Khatri ChhetriDocument49 pagesAutomobile Tracking System Using Gps and GSM: Jaya Ram Khatri Chhetrioluwakayode olabanjiNo ratings yet
- Siminar SajaDocument29 pagesSiminar SajaMarwan CompNo ratings yet
- RJP30H2ADocument6 pagesRJP30H2AEdgar OcantoNo ratings yet
- Saurab Gurung 20MC204001 Research MethodologyDocument36 pagesSaurab Gurung 20MC204001 Research Methodologyyogendra sharmaNo ratings yet
- Python Hands On AnswersDocument15 pagesPython Hands On AnswersSai GopiNo ratings yet
- PiLog Group Profile MES EXTDocument21 pagesPiLog Group Profile MES EXTAdil GibreelNo ratings yet
- Slide 2: Early Life SuccessDocument4 pagesSlide 2: Early Life SuccessSandhya SharmaNo ratings yet
- Finite State MachinesDocument17 pagesFinite State Machineslashali1212No ratings yet
- SCWCD QuestionsDocument18 pagesSCWCD QuestionsRitesh JhaNo ratings yet
- Java Beans and JSP ActionsDocument55 pagesJava Beans and JSP Actionsaaaaaaa2010No ratings yet
- NotebudgetDocument2 pagesNotebudgetandrei santosNo ratings yet
- UiPath Tutorial - JavatpointDocument35 pagesUiPath Tutorial - Javatpointkarthy143No ratings yet
- ThinkScript Strategy With PAR SARDocument4 pagesThinkScript Strategy With PAR SARbollingerbandNo ratings yet
- Roland Kc100Document8 pagesRoland Kc100J SusiloNo ratings yet
- Leaflet For Paper Presentation G P Arvi 31-01-24. UpdatedDocument2 pagesLeaflet For Paper Presentation G P Arvi 31-01-24. UpdatedAditya AnandNo ratings yet
- Steady-State Model and Power-Flow Analysis of Single-Phase Electronically Coupled Distributed Energy ResourcesDocument9 pagesSteady-State Model and Power-Flow Analysis of Single-Phase Electronically Coupled Distributed Energy Resources॰अचूक॰ Milan SubediNo ratings yet
- Enterasys Nac GuideDocument19 pagesEnterasys Nac GuideIchwan SetiawanNo ratings yet
- HMI Configuration Setup r1Document13 pagesHMI Configuration Setup r1Mohamed AmineNo ratings yet
- Long Beach, A Sunlife Resort - 5 Star Luxury Resort in MauritiussDocument1 pageLong Beach, A Sunlife Resort - 5 Star Luxury Resort in MauritiussdsfgfNo ratings yet