Professional Documents
Culture Documents
Comparision of SCANRDI Vs Pharmacopiea Method
Comparision of SCANRDI Vs Pharmacopiea Method
Uploaded by
Umair ShekhaniCopyright:
Available Formats
You might also like
- SOP For Clinical PharmacyDocument64 pagesSOP For Clinical PharmacyRaymond Ofori100% (3)
- Biomerieux ScanRDI Manual - EnglishDocument91 pagesBiomerieux ScanRDI Manual - Englishblack betty100% (1)
- Rapid Microbiology Method by Jeane MoldenhauerDocument24 pagesRapid Microbiology Method by Jeane MoldenhauerMaurits TobingNo ratings yet
- Upsteam ProcessingDocument21 pagesUpsteam ProcessingAdithyaNo ratings yet
- Sutton The Sterility TestsDocument26 pagesSutton The Sterility TestsSilke IgemannNo ratings yet
- Rapid Microbiological Methods They Are Rapid Are They FastDocument9 pagesRapid Microbiological Methods They Are Rapid Are They FastRonald SalasNo ratings yet
- Methods of Rapid Microbiological AssayDocument10 pagesMethods of Rapid Microbiological AssayMundi OdiumNo ratings yet
- En Do Toxin CatalogDocument17 pagesEn Do Toxin CatalogmarronyyyNo ratings yet
- Biologic License ApplicationDocument16 pagesBiologic License ApplicationJean Sandra PintoNo ratings yet
- The Future of Monoclonal AntibodyDocument180 pagesThe Future of Monoclonal AntibodyJohn John100% (1)
- ANDA's Impurities in Drug SubstancesDocument19 pagesANDA's Impurities in Drug SubstancesparagnkpatelNo ratings yet
- Use and Limitations of in Vitro Dissolution Testing: Topic Introduction and OverviewDocument114 pagesUse and Limitations of in Vitro Dissolution Testing: Topic Introduction and OverviewMuthu Venkatesh100% (1)
- CMCDocument3 pagesCMCSri harsha reddyNo ratings yet
- Microbiological Pharmaceutical Quality Control Labs (7 - 93) - FDADocument8 pagesMicrobiological Pharmaceutical Quality Control Labs (7 - 93) - FDAArmando SaldañaNo ratings yet
- SOP For Antimicrobial Effectiveness TestingDocument4 pagesSOP For Antimicrobial Effectiveness TestingGencay ErginNo ratings yet
- Pda: A Global Association: Recombinant Factor C - EndolisaDocument29 pagesPda: A Global Association: Recombinant Factor C - EndolisaPiruzi MaghlakelidzeNo ratings yet
- 5 1 10 Guidelines For Using The Test For Bacterial EndotoxinsDocument5 pages5 1 10 Guidelines For Using The Test For Bacterial Endotoxinsnguyentuanson167100% (1)
- FDA Guidance For Industry S7a Safety Pharmacology Studies For Human Pharmaceuticals PDFDocument14 pagesFDA Guidance For Industry S7a Safety Pharmacology Studies For Human Pharmaceuticals PDFbmartindoyle6396No ratings yet
- 1.1 Analytical Chemistry: Introductio NDocument56 pages1.1 Analytical Chemistry: Introductio NmadhubaddapuriNo ratings yet
- FDA Guidance For CMC For Clinical TrialsDocument27 pagesFDA Guidance For CMC For Clinical TrialsNelson Alejandro FierroNo ratings yet
- Ophthalmic Drug Delivery Systems, 2ed. 2003Document727 pagesOphthalmic Drug Delivery Systems, 2ed. 2003VuqarNo ratings yet
- Stability of Pharmaceuticals-1Document96 pagesStability of Pharmaceuticals-1Hely PatelNo ratings yet
- ObjectionableDocument9 pagesObjectionabledmtalbhogeNo ratings yet
- Sterile Parenteral Product (Review) PDFDocument8 pagesSterile Parenteral Product (Review) PDFdianiNo ratings yet
- FDA - ICH M7 (R1) - Control of Mutagenic Impurities in Pharmaceuticals 03.2018Document131 pagesFDA - ICH M7 (R1) - Control of Mutagenic Impurities in Pharmaceuticals 03.2018Catrinescu OanaNo ratings yet
- The New Concept of Automatic Gloved Hand SanitizationDocument3 pagesThe New Concept of Automatic Gloved Hand SanitizationTim SandleNo ratings yet
- CTD CMC-Information-Human-Gene-Therapy-IND-Applications - Jan - 2020Document56 pagesCTD CMC-Information-Human-Gene-Therapy-IND-Applications - Jan - 2020Shiraz KhanNo ratings yet
- Practical Guide To Autoclave Validation: by Raymond G. Lewis, PEDocument8 pagesPractical Guide To Autoclave Validation: by Raymond G. Lewis, PEManarKhNo ratings yet
- Guidelines For Cell LinesDocument26 pagesGuidelines For Cell LinesexecNo ratings yet
- Antimicrobial EffectivenessDocument6 pagesAntimicrobial EffectivenessChristineNo ratings yet
- Anti Microbiological Assay Test or Antibiotic Assay Test of Pharmaceutical Preparation Containing Antibiotics Using 'Cylinder Plate Method'Document4 pagesAnti Microbiological Assay Test or Antibiotic Assay Test of Pharmaceutical Preparation Containing Antibiotics Using 'Cylinder Plate Method'Editor IJTSRDNo ratings yet
- Microbial Contamination-A Regulatory Perspective.Document8 pagesMicrobial Contamination-A Regulatory Perspective.Sameer PanditNo ratings yet
- Risk Assessment in The Pharmaceutical Industry ToxicologyDocument41 pagesRisk Assessment in The Pharmaceutical Industry ToxicologyJasonNo ratings yet
- Usp 601Document33 pagesUsp 601Nur AcarNo ratings yet
- Successful Sterility Test Failure Investigations-A Practical ApproachDocument5 pagesSuccessful Sterility Test Failure Investigations-A Practical ApproachMiniq ForondaNo ratings yet
- Mammalian Expression SystemsDocument16 pagesMammalian Expression SystemsfdjkafjkljdklfjalNo ratings yet
- Valiadasi Method CleainhgDocument76 pagesValiadasi Method CleainhgYulis AdrianaNo ratings yet
- Seminar (Photostability)Document12 pagesSeminar (Photostability)Mr. HIMANSHU PALIWALNo ratings yet
- Fda 1571Document2 pagesFda 1571Nitin KashyapNo ratings yet
- Sterile Ophthalmic Preparations and Cont PDFDocument7 pagesSterile Ophthalmic Preparations and Cont PDFRaedMohNo ratings yet
- 62 The Basics of Bioburden TestingDocument2 pages62 The Basics of Bioburden Testinghitham shehataNo ratings yet
- Mark Old Corne Microbiological ValidationDocument28 pagesMark Old Corne Microbiological ValidationhumusdelombrizNo ratings yet
- Cundell Et Al PDA J Pharm Sci Tech March 2010 (Full Article) SfsDocument21 pagesCundell Et Al PDA J Pharm Sci Tech March 2010 (Full Article) SfsJessica González LaraNo ratings yet
- Gamma IrradiationDocument27 pagesGamma Irradiationmuzammil21_adNo ratings yet
- 1111 - USP Micro Limit Test For Non SterileDocument2 pages1111 - USP Micro Limit Test For Non SterileSpectre Spectre100% (1)
- Patent-US 20070003628 A1-Nanoparticulate Clopidogrel FormulationsDocument19 pagesPatent-US 20070003628 A1-Nanoparticulate Clopidogrel FormulationsQuan NguyenNo ratings yet
- PUPSITDocument43 pagesPUPSITAlessandro ChreimNo ratings yet
- PQLI Definition of CriticalityDocument10 pagesPQLI Definition of CriticalitypakdekroNo ratings yet
- 23635.1.5. Application of The F0 Concept To Steam Sterilisation of Aqueou - PDFDocument1 page23635.1.5. Application of The F0 Concept To Steam Sterilisation of Aqueou - PDFlilaNo ratings yet
- Quality Control and Drug AnalysisDocument57 pagesQuality Control and Drug AnalysisIhab AdelNo ratings yet
- SFDA Saudi Biosimilar GuidelinesDocument17 pagesSFDA Saudi Biosimilar Guidelinesshivani hiremath100% (1)
- Preventing Metal ContaminationDocument6 pagesPreventing Metal Contaminationlouish9175841No ratings yet
- Development of New Ophthalmic Suspension Prednisolone AcetateDocument6 pagesDevelopment of New Ophthalmic Suspension Prednisolone AcetaterandatagNo ratings yet
- Old Drugs For A New Use (Prescription)Document17 pagesOld Drugs For A New Use (Prescription)Karol Denisse Hernández BastidaNo ratings yet
- Selection of DissolutionDocument5 pagesSelection of DissolutionGirishNo ratings yet
- USP Seminar - Fundamentals of Bioassay Practices 2014 PDFDocument140 pagesUSP Seminar - Fundamentals of Bioassay Practices 2014 PDFnsk79in@gmail.comNo ratings yet
- Us P 797 Pharmaceutical Compounding Sterile CompoundingDocument61 pagesUs P 797 Pharmaceutical Compounding Sterile Compoundingdeepa_ragu5695No ratings yet
- Burkholderia CepaciaDocument11 pagesBurkholderia Cepaciaportner6873No ratings yet
- Container Closure IntegrityDocument19 pagesContainer Closure IntegrityYessieNo ratings yet
- A Comprehensive Text Book on Self-emulsifying Drug Delivery SystemsFrom EverandA Comprehensive Text Book on Self-emulsifying Drug Delivery SystemsNo ratings yet
- Enzyme Inhibition in Drug Discovery and Development: The Good and the BadFrom EverandEnzyme Inhibition in Drug Discovery and Development: The Good and the BadChuang LuNo ratings yet
- Exercise Cost of CapitalDocument7 pagesExercise Cost of CapitalUmair Shekhani100% (2)
- Exercise Capital BudgetingDocument2 pagesExercise Capital BudgetingUmair ShekhaniNo ratings yet
- Exercise Capital BudgetingDocument2 pagesExercise Capital BudgetingUmair ShekhaniNo ratings yet
- Balanced ScorecardDocument56 pagesBalanced ScorecardUmair ShekhaniNo ratings yet
- RD of CBD ProductsDocument13 pagesRD of CBD ProductsUmair ShekhaniNo ratings yet
- Assignment # 2: Book: From Financial Accounting, Ninth Edition, by Weygandt, Kimmel, KiesoDocument1 pageAssignment # 2: Book: From Financial Accounting, Ninth Edition, by Weygandt, Kimmel, KiesoUmair ShekhaniNo ratings yet
- Business CommunicationDocument18 pagesBusiness CommunicationUmair ShekhaniNo ratings yet
- Exercise Stock ValuationDocument2 pagesExercise Stock ValuationUmair ShekhaniNo ratings yet
- Commercial Facility For CBD ProductsDocument6 pagesCommercial Facility For CBD ProductsUmair ShekhaniNo ratings yet
- A. Introduction / Background: Industrial and Medicinal Management of Hemp As An Agriculture Commodity by Indus GroupDocument4 pagesA. Introduction / Background: Industrial and Medicinal Management of Hemp As An Agriculture Commodity by Indus GroupUmair ShekhaniNo ratings yet
- Adr Form 1Document2 pagesAdr Form 1Umair ShekhaniNo ratings yet
- Adr Form 2Document2 pagesAdr Form 2Umair ShekhaniNo ratings yet
- Indus Pharma (PVT.) LTD.: Protocol For Re-Qualification of Water Distillation Plant (Wfi), Storage & Distribution SystemDocument9 pagesIndus Pharma (PVT.) LTD.: Protocol For Re-Qualification of Water Distillation Plant (Wfi), Storage & Distribution SystemUmair Shekhani100% (1)
- Inspection Report of Incoming PMDocument1 pageInspection Report of Incoming PMUmair ShekhaniNo ratings yet
- Standard Operating Procedure: Indus Pharma (PVT.) LTDDocument4 pagesStandard Operating Procedure: Indus Pharma (PVT.) LTDUmair ShekhaniNo ratings yet
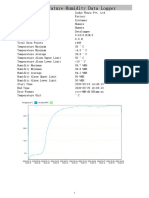
- Temperature Mapping DataDocument13 pagesTemperature Mapping DataUmair ShekhaniNo ratings yet
- Standard Operating Procedure: Indus Pharma (PVT.) LTDDocument4 pagesStandard Operating Procedure: Indus Pharma (PVT.) LTDUmair ShekhaniNo ratings yet
- Indus Pharma (PVT.) LTD.: Complaint Response FormDocument1 pageIndus Pharma (PVT.) LTD.: Complaint Response FormUmair ShekhaniNo ratings yet
- EMA Epar Public Assessment ReportDocument37 pagesEMA Epar Public Assessment ReportOscar Javier EstupiñánNo ratings yet
- Formula Ambroxol JurnalDocument3 pagesFormula Ambroxol Jurnalrd_al_snrNo ratings yet
- Summer Training Report AbhiDocument51 pagesSummer Training Report AbhiAbhijit MohantyNo ratings yet
- Mimusops Elengi Linn:a ReviewDocument9 pagesMimusops Elengi Linn:a ReviewRAPPORTS DE PHARMACIENo ratings yet
- Usp 36-NF 31 - GC41Document1 pageUsp 36-NF 31 - GC41DennyNo ratings yet
- Why Companies Are Selling Same Drug in Different PricesDocument2 pagesWhy Companies Are Selling Same Drug in Different Pricesdharmendra gaurNo ratings yet
- Pharmacist Syllabus DetailedDocument18 pagesPharmacist Syllabus DetailedHari Kamesh KiranNo ratings yet
- HhihhygtfrtdgbhjDocument11 pagesHhihhygtfrtdgbhjKania HidayantiNo ratings yet
- GCC Data Requirements For Human Drugs Submission Version 1.1 PDFDocument81 pagesGCC Data Requirements For Human Drugs Submission Version 1.1 PDFBasha Yazn Anjak50% (2)
- uk-spc-nexium-sachet-10mg-DRESS-update-GI 21 0012aDocument17 pagesuk-spc-nexium-sachet-10mg-DRESS-update-GI 21 0012aDeisy ClerkeNo ratings yet
- Jurnal LUTS PDFDocument8 pagesJurnal LUTS PDFmitaNo ratings yet
- Deflazen: Training ManualDocument10 pagesDeflazen: Training Manualanupdr_cNo ratings yet
- Medco Express FINALDocument23 pagesMedco Express FINALSean MooreNo ratings yet
- Three Different Types of Airlocks in Pharmaceuticals - Pharma PathwayDocument4 pagesThree Different Types of Airlocks in Pharmaceuticals - Pharma PathwayFaizul ZainudinNo ratings yet
- ExperimentDocument9 pagesExperimentsatyakrishna1234No ratings yet
- List of Cos in AhmedabadDocument559 pagesList of Cos in AhmedabadVikhyat SharmaNo ratings yet
- Drug Development Process - Part 1Document5 pagesDrug Development Process - Part 1Nguyễn Thế ThaoNo ratings yet
- Pharm MCQDocument17 pagesPharm MCQArun Kumar100% (2)
- Cough Syrup Detail:: Prodactive or Expectorant and Suppressant or Dry CoughDocument3 pagesCough Syrup Detail:: Prodactive or Expectorant and Suppressant or Dry CoughFarhadullah KhanNo ratings yet
- Barros 2014 - The Use of Pictograms in The Health CareDocument16 pagesBarros 2014 - The Use of Pictograms in The Health CareBlaberNo ratings yet
- Drug Delivery SystemsDocument8 pagesDrug Delivery SystemsNikko Nabasca GorneNo ratings yet
- List of Abbreviations Used in Medical PrescriptionsDocument5 pagesList of Abbreviations Used in Medical PrescriptionsNathan TonthatNo ratings yet
- Lista de Precios Farmacos 27-04-2022Document5 pagesLista de Precios Farmacos 27-04-2022Mery Carolina PerezNo ratings yet
- The Science of Ischemic Stroke Pathophysiology Pharmacological TreatmentDocument20 pagesThe Science of Ischemic Stroke Pathophysiology Pharmacological TreatmentLisa Linggi'AlloNo ratings yet
- Switching P2Y Inhibitors: Medication SafetyDocument1 pageSwitching P2Y Inhibitors: Medication SafetyWaleed saleemNo ratings yet
- Presented By: Samiksha B.Sawant M, PHARM (IP), 1 SEM Guided By: Dr. Indira ParabDocument46 pagesPresented By: Samiksha B.Sawant M, PHARM (IP), 1 SEM Guided By: Dr. Indira ParabKainat MalikNo ratings yet
- Pharmacy Daily For Wed 09 Sep 2015 - Australia-China Drug Pact, Pharmacy Continence Courses, TWC Hearing Tests, Health AMPERSAND Beauty and Much MoreDocument4 pagesPharmacy Daily For Wed 09 Sep 2015 - Australia-China Drug Pact, Pharmacy Continence Courses, TWC Hearing Tests, Health AMPERSAND Beauty and Much MorepharmacydailyNo ratings yet
- Friends PharmaDocument3 pagesFriends PharmaaoryaNo ratings yet
- 70-Article Text-286-2-10-20221205Document7 pages70-Article Text-286-2-10-20221205Nurul Maulida PutriNo ratings yet
Comparision of SCANRDI Vs Pharmacopiea Method
Comparision of SCANRDI Vs Pharmacopiea Method
Uploaded by
Umair ShekhaniCopyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Comparision of SCANRDI Vs Pharmacopiea Method
Comparision of SCANRDI Vs Pharmacopiea Method
Uploaded by
Umair ShekhaniCopyright:
Available Formats
Downloaded from journal.pda.
org on September 1, 2010
Evaluation of the ScanRDI® as a Rapid Alternative to the
Pharmacopoeial Sterility Test Method: Comparison of the
Limits of Detection
Ron Smith, Mark Von Tress, Cheyenne Tubb, et al.
PDA J Pharm Sci and Tech 2010, 64 356-363
Downloaded from journal.pda.org on September 1, 2010
Evaluation of the ScanRDI姞 as a Rapid Alternative to the
Pharmacopoeial Sterility Test Method: Comparison of the
Limits of Detection
RON SMITH, MARK VON TRESS, CHEYENNE TUBB, and ERWIN VANHAECKE
Alcon Laboratories Inc., 6201 S. Freeway, Fort Worth, Texas 76134 ©PDA, Inc. 2010
ABSTRACT: Two sterility test methods, the ScanRDI威 rapid sterility test and the United States Pharmacopeia/
European Pharmacopoeia/Japanese Pharmacopoeia (USP/EP/JP) compendial sterility test, were compared with
respect to the limits of detection for the presence of viable microorganisms in aqueous solutions at low inoculation
levels. The ScanRDI威 system employs a combination of direct fluorescent labeling techniques and solid-phase laser
scanning cytometry to rapidly enumerate viable microorganisms from aqueous samples, whereas the compendial
sterility test is a qualitative, growth-based method that uses a visual assessment of turbidity to indicate microbial
contamination. Eight microorganisms were evaluated, seven compendial microorganisms (Clostridium sporogenes,
Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Bacillus subtilis, Aspergillus niger, Candida
albicans) and the Gram-positive anaerobe Propionibacterium acnes. The number of viable organisms was estimated
using the ScanRDI威 method and the conventional sterility test method using most probable number methodology. The
mean difference between the methods was computed and 95% confidence intervals around the mean difference were
estimated. The ScanRDI威 method was found to be numerically superior and statistically non-inferior to the
compendial (USP/EP/JP) sterility test with respect to the limits of detection for all organisms tested.
KEYWORDS: Rapid sterility test, Chemunex ScanRDI威, Most probable number, Sterility testing
Introduction A sterility test, in its most basic form, is a qualitative
assay that is designed to detect the absence of viable
A common experimental approach to demonstrating microbial cells in or on a product. The expected out-
functional equivalence between two analytical meth- come for such a test is zero, or no organisms recov-
ods is to conduct a series of parallel, or side-by-side, ered. In the current pharmaceutical environment, the
experiments. Such studies are best suited to those likelihood of observing a sterility failure is exceed-
quantitative microbiological methods that characterize ingly rare, generally on the order of less than 0.1%. As
populations. In these methods, the abundance of mi- such, studies designed to demonstrate functional
crobial cells allows for statistical evaluation of the equivalence for these types of methods, using the
experimental outcomes. However, the parallel testing qualitative presence/absence metric, will require inor-
approach is not well suited to compare methods that dinately large sample sizes, which become signifi-
are qualitative in nature, or those that attempt to detect cantly larger as the differences between two methods
and quantify rare events (i.e., detecting microbial con- shrink (1).
tamination at extraordinarily low concentrations).
With these methods, the inherent statistical power of An alternate approach to evaluating equivalence be-
the microbial population is lost, and it must therefore tween such qualitative methods is through the appli-
be provided through increased repetition of the exper- cation of the most probable number (MPN) method.
iment. The MPN method is well-known and widely used for
estimating the density of viable microorganisms based
on the Poisson distribution (2, 3). A sample is diluted
to such an extent that only a small proportion of
sub-samples will contain organisms that will display
Received July 14, 2009.
positive growth when assayed. It is thus possible to
Please address correspondence to Ron Smith. Phone:
use a series of qualitative assays, such as a sterility
817.551.4975. e-mail: Ron.Smith2@alconlabs.com
test, in an MPN series, to provide a quantitative mea-
356 PDA Journal of Pharmaceutical Science and Technology
Downloaded from journal.pda.org on September 1, 2010
surement of the number of viable organisms in a filter is scanned by a high-speed, 488-nm argon laser.
solution. More importantly, the MPN method provides The entire filter surface is scanned in a pattern that
a means to utilize a quantitative non-inferiority crite- ensures that each section of the membrane is illumi-
rion for a qualitative test method. Non-inferiority stud- nated twice. Fluorescent light is detected by multiple
ies are designed to show that a particular test method photomultiplier tubes and processed through a number
is no worse than a standard method by a given margin. of discrimination parameters that enable the instru-
Non-inferiority testing further permits the conclusion ment to differentiate between valid signals (labeled
of superiority. microbes) and background noise (electronic, optical,
and/or auto-fluorescent particles). The result of the
In theory, the presence of a single viable cell should be scan is displayed in the form of a membrane scan map
sufficient to generate a positive result in a sterility test. that identifies the position of each fluorescent event.
Consequently, the limit of detection is the single most Each event is verified by visual examination using an
important attribute of a sterility test method. In this epifluorescent microscope with an automated motor-
study we employed an MPN scheme to compare the ized stage. Those events that possess morphology con-
ability of two sterility test methods to detect microor- sistent with microbial cells are subsequently consid-
ganisms when they were exceedingly rare. We com- ered viable microorganisms. Amorphous particles and
pared a rapid detection approach using the ScanRDI威 auto-fluorescing crystals are invalidated. The com-
microbial analysis system and the reference sterility bined speed and sensitivity of the ScanRDI威 system
test method described in the current US/EP/JP phar- make it ideally suited for industrial applications, par-
macopeias (United States Pharmacopeia/European ticularly for ascertaining the sterility of filterable so-
Pharmacopoeia/Japanese Pharmacopoeia). We also lutions (4 – 6).
evaluated the robustness of the methods by replicating
the experiments using three discrete teams of analysts Sterility Test Methods
in three different laboratories. The data generated
from these studies support the conclusion that the Organisms and Culture Conditions: Organisms,
ScanRDI威 sterility test method is non-inferior to the ATCC identification, and growth conditions used in
compendial sterility test with respect to the limits of each MPN series are given in Table I. Organisms, with
detection. the exception of Propionbacterium acnes, were ob-
tained as lyophilized Bioballs™ from BTF Precise
Materials and Methods Microbiology (Pittsburgh, PA). Bacillus subtilis, Clos-
tridium sporogenes, and Aspergillus niger were ob-
Overview of the ScanRDI威 System tained as preparations of spores, while the remaining
organisms were vegetative cells. Bioballs were dis-
The ScanRDI威 microbial analysis system employs a solved in sterile saline to yield an initial concentration
combination of direct fluorescent labeling and solid- of 30 cfu/mL⫺1. Subsequent dilutions were performed
phase laser scanning cytometry to rapidly enumerate in saline to yield three dilutions: 10⫺1, 10⫺2, and
viable microorganisms from aqueous samples. The 10⫺3. P. acnes was serially diluted in sterile saline to
system has sufficient sensitivity to detect a single yield cell concentrations ranging between 0.03 and 3
viable microorganism in 2–3 h, without the need for cfu/mL⫺1. Culture conditions (media, temperature)
growth and multiplication. Cells are collected by fil- were based on the compendial requirements for steril-
tration onto a single, 0.4-m-porosity polyester mem- ity testing and are indicated in Table I.
brane and treated with a proprietary combination of
background and viability stains. The viability stain ScanRDI威 Test Method: At each dilution, the con-
consists of a membrane-permeant, non-fluorescent tents of a tube were briefly vortexed and a 1-mL
substrate, similar to fluorescein diacetate, which freely aliquot was filtered through a fluorassure integrated
crosses the cell membrane. The substrate is cleaved by filtration unit (FIFU) (Chemunex 220-C2104-02). The
nonspecific esterases into a membrane-impermeant filter was incubated aerobically on resuscitation media
chromophore. As a result, cells with intact membranes for 3.0 h ⫾ 10 min at 30 –35 °C prior to labeling with
accumulate the chromophore in the cytoplasm while the viability reagent. This resuscitation step is re-
those with compromised membranes are unable to quired to activate the metabolic activity of dormant
retain the fluorescent probe. A processed filter is sub- spores and to induce germination such that the spores
sequently transferred into the cytometer where the can be labeled with the viability reagent (6, 7, 9).
Vol. 64, No. 4, July–August 2010 357
Downloaded from journal.pda.org on September 1, 2010
TABLE I
Microorganisms and Culture Conditions
Incubation
Strain Growth Temperature
Organism Numbera Mediumb (°C)
Clostridium sporogenes 11437 FTM 30–35
Propionibacterium acnes 6919 FTM 30–35
Escherichia coli 8739 FTM 30–35
Pseudomonas aeruginosa 9027 FTM 30–35
Staphylococcus aureus 6538 FTM 30–35
Bacillus subtilis 6633 TSB 20–25
Aspergillus niger 16404 TSB 20–25
Candida albicans 10231 TSB 20–25
a
American Type Culture Collection.
b
FTM ⫽ fluid thioglycollate medium; TSB ⫽ tryptic soy broth.
Under these conditions both aerobic and anaerobic assay. For enumeration, a 1-mL aliquot from each
bacteria spores as well as mold spores can be quanti- dilution tube in the MPN series was directly inocu-
tatively detected by the ScanRDI威 system with no lated into 100 mL of the appropriate culture medium
effect on the vegetative forms. While longer incuba- and incubated for 14 days at the recommended tem-
tion periods may improve the recovery and detection perature (Table I). Positive growth was determined by
of spores, extended periods of incubation adversely a visual assessment of turbidity.
affect the ability to efficiently label and detect other
microorganisms, particularly those that have been Quantitation by MPN
physically damaged or subjected to metabolic and
environmental stressors. As such, the parameters (time To create the MPN series, a suspension of each mi-
and temperature) of the pre-labeling step were opti- croorganism was serially diluted, as described previ-
mized to achieve accurate recovery of all the test ously, to yield suspensions containing approximately
organisms, and by inference, the broadest spectrum of 3.0, 0.3, and 0.03 cfu/mL. From each dilution tube, a
organisms possible. Following the pre-labeling step, series of 1-mL aliquots was aseptically transferred into
the filters were aseptically transferred onto an absor- each of five FIFU assemblies, and similarly into each
bent pad saturated with the proprietary fluorogenic of five culture vessels, thus creating two 5-tube MPN
stain (Chemunex 201-R2007-03) and incubated series (see http://www.cfsan.fda.gov/⬃ebam/bam-
(45– 60 min at 30 –35 °C, aerobic conditions). The a2.html) (12). The concentration of microorganisms in
processed filters were transferred into the cytometer one MPN series (FIFU) was determined using the
and scanned using discrimination parameters deter- ScanRDI威 method, while estimates of the microorgan-
mined by the manufacturer. Each fluorescent event ism concentrations in the other series were determined
was verified by visual examination using an epifluo- using the reference sterility test method.
rescent microscope with an automated motorized stage
at a magnification of 500⫻. Those events morpholog- Statistical Methods
ically consistent with microbial cells were considered
viable microorganisms and enumerated by the analyst. Test Method and Early Stopping Rule: A paired t-test
For the purpose of this study, the presence of one or was used to test the hypothesis that the concentration
more validated fluorescent events was scored as a of microorganisms detected by ScanRDI威 is less than
positive sterility result. or equal to 70% of the concentration of microor-
ganisms detected and enumerated by the conventional
Direct Sterility Test Method: Test solutions were sterility test method (i.e., the ([ScanRDI威 mean
assessed for sterility using a direct inoculation test log10(MPN)] ⫺ [Conventional mean log10(MPN)]) ⬎
method (9 –11). This increased assurance that the ap- (log10(0.7))) is not inferior to that of the compendial
propriate levels of organisms were delivered into the sterility test method) (13). The experimental designed
358 PDA Journal of Pharmaceutical Science and Technology
Downloaded from journal.pda.org on September 1, 2010
allowed up to 25 pairs of MPN determinations to be Robustness Evaluation: Experiments were replicated
collected for each organism. A confidence limit of in three locations (Alcon Research and Development
95.25% is required to control for an overall type I Lab in Fort Worth, Texas; Alcon Quality Assurance
error using this number of samples. Consequently, a Lab in Fort Worth, Texas; and Alcon Quality Assur-
test of the hypothesis was performed by comparing a ance Lab in Kaysersberg, France; 120 tests per site)
lower, one-sided 95.25% confidence limit on the with three teams of analysts. An analysis of variance
paired difference (log-transformed [log10] bacterial was used to estimate site-to-site variability. The model
concentration determined from the MPN series using is given by
ScanRDI威 minus the log-transformed concentration of
bacteria determined using the reference sterility test Y l共ijk兲 ⫽ ⫹ T i ⫹ ␥ j ⫹ ␦ k共 j兲 ⫹ ε l共ijk兲
method) to the index of non-inferiority (⫺0.1549 ⫽
log10(0.7)). where Y l(ijk) is a measurement of log MPN, is an
intercept, T i is the fixed treatment effect, ␥ j is a
Fewer than 25 pairs of samples for each organism random effect for sites, ␦ k( j) is a random effect for
were collected if there was at least a 95% probability treatments within sites, and ⑀ l(ijk) is a random effect
of rejecting the hypothesis using fewer samples; how- for the individual measurements of log10 (MPN). The
ever, the minimum sample size was 10. An early random effect for sites, ␥ j , is assumed to be normally
stopping rule was established using the test statistic z n distributed with a mean of zero and a variance for sites
and critical values determined according to Jennison of ␥2 , which is tested to be zero. If the site-to-site
and Turnbull (14 [see p 207, eq 10.3]). The test variance is not significantly greater than zero, then the
statistic was calculated according to statistical inference may be considered robust to vari-
ation among sites.
共d ⫹ 0.1549兲 冑n
zn ⫽ Results
sd
The results presented in this report indicate that the
where d is the difference in the means between the ScanRDI威 method is numerically superior and statis-
log-transformed (log10) bacterial concentration deter- tically non-inferior to the reference sterility test
mined from the MPN series using ScanRDI威 minus method with respect to the limits of detection for all of
the log-transformed concentration of bacteria deter- organisms evaluated (Table II). The differences be-
mined using the reference sterility test method, n is the tween mean MPN values were always positive, indi-
number of samples, and s d is the standard deviation of cating that the ScanRDI威 method was consistently
the paired differences. better at detecting viable organisms at exceedingly
low concentrations (Figure 1). However, the confi-
Determination of Limits of Detection: The limit of dence interval around the mean difference for Clos-
detection was the dilution level at which there is tridium sporogenes includes zero, which indicates that
97.5% confidence that there was less than 10% chance both methods are at least equivalent in their ability to
of detecting organisms in a sample. The 97.5 % con- detect this organism. Since the lower one-sided
fidence level stems from the upper limit of a two-sided 95.25% confidence limit on the paired difference for
95% confidence interval for the probability of detect- each organism was greater than ⫺0.1549, the accep-
ing an organism. The limit of detection was deter- tance criteria were satisfied and the non-inferiority
mined from repeated measures logistic regression us- null hypothesis of inferiority was rejected. Addition-
ing three dilutions (as above), five tubes per dilution, ally, the robustness of the ScanRDI威 method was
and the number of tubes indicating the presence of assessed using analysis of variance to estimate the
microorganisms. The predictor variable was the log10 site-to-site variability and determine if it was signifi-
of the dilution. Four thousand two-sided 95% confi- cantly different from zero. The site-to-site variance
dence limits on the predicted probability of response component in this analysis measures the method’s
were computed, and the log value producing the larg- consistency among laboratories. The results of this
est upper confidence limit less than or equal to 0.1 analysis are presented in Table III and indicate that the
(10%) was selected as the limit of detection. The data ScanRDI威 sterility test method remains superior or
analysis was generated using SAS/STAT software, equivalent to the reference sterility method in the
Version 9.1 of the SAS System for Windows (15). determination of mean MPN after adjusting for vari-
Vol. 64, No. 4, July–August 2010 359
Downloaded from journal.pda.org on September 1, 2010
TABLE II
Paired T-Tests of Non-Inferiority of the ScanRDI威 and Compendial Sterility Test
Difference Lower
between Standard Confidence
Organism Mean MPNa Deviation n zn P Limitb
Clostridium sporogenes 0.05 0.29 24 3.518 0.0009 ⫺0.050
Propionibacterium acnes 0.58 0.40 15 7.128 ⬍0.0001 0.398
Escherichia coli 0.20 0.30 25 5.812 ⬍0.0001 0.092
Pseudomonas aeruginosa 0.95 0.37 13 10.972 ⬍0.0001 0.762
Staphylococcus aureus 0.39 0.29 14 7.067 ⬍0.0001 0.251
Bacillus subtilis 0.43 0.23 12 8.904 ⬍0.0001 0.308
Aspergillus niger 0.67 0.41 16 8.115 ⬍0.0001 0.490
Candida albicans 0.64 0.46 13 6.278 ⬍0.0001 0.412
a
Mean log10 MPN for ScanRDI威 minus mean log10 MPN for compendial sterility method.
b
The acceptance criteria were satisfied if the lower one-sided 95.25% confidence limit on the paired difference was
less than ⫺0.1549, which is log10(0.70).
ation among sites. Moreover, the site-to-site variabil- Because the study was designed to estimate MPN, the
ity in MPN measurement is not significantly different data could not support a comparison of accuracy,
from zero for any organism tested in the limit of precision, and specificity between methods, nor could
detection study. a false-positive rate be established. However, the data
indicate that the overall variability (as range in percent
positives) of the reference sterility test method ap-
peared to be greater than that of the ScanRDI威 method
when microbes were inoculated into an MPN series at
a cell concentration of 3 cells/mL⫺1. The overall vari-
ability of the reference method appeared to decrease
when cells were present at lower concentrations (0.03
cells/mL⫺1). However, this may be misleading be-
cause the reference method rarely recovered bacteria
at low cell abundance (high frequency of zero values)
but cells were more often recovered by the ScanRDI威
(Figure 2, percent positives greater than zero). For
example, when all eight organisms were considered,
the percentage of positive MPN tubes containing 0.03
bacteria/mL⫺1 was 2.12% for the reference method
but 17.88% for the ScanRDI威. The difference in the
ability of the two methods to recover bacteria is
clearly revealed in Figure 2. Not only was the
ScanRDI威 better able to reveal the presence of bacte-
Figure 1 ria when they were very rare, analysis of the data
indicate that the limit of detection of the ScanRDI威 is
Difference between mean concentrations of bacte- about an order of magnitude lower than that of the
ria detected by ScanRDI威 and the compendial reference sterility test method (Table IV).
methods for each of eight bacteria. Data points
falling within the shaded region would indicate that Discussion
the ScanRDI威 method was inferior to the compen-
dial sterility test method for detecting contami- One of the challenges for any sterility test is the ability
nants. Error bars represent the two 1-sided 95% to detect or recover contamination, even when con-
confidence intervals. taminants are exceedingly rare. In this study we es-
360 PDA Journal of Pharmaceutical Science and Technology
Downloaded from journal.pda.org on September 1, 2010
TABLE III
Analysis of Variance for Robustness Evaluation
One-Sided
Lower 95.5%
Confidence
Limit Site
Organism Log10(MPN)a Variance P > 円Z円b
Clostridium sporogenes ⫺0.050 0.0093 0.2522
Propionibacterium acnes 0.415 0.0857 0.1780
Escherichia coli 0.092 0.0000 1.0000
Pseudomonas aeruginosa 0.771 0.0218 0.2366
Staphylococcus aureus 0.264 0.0138 0.2227
Bacillus subtilis 0.308 0.0054 0.3167
Aspergillus niger 0.490 0.0014 0.4544
Candida albicans 0.438 0.0270 0.2392
a
The confidence coefficient selection reflects a 0.5% adjustment for interim analysis. The lower confidence limits are
slightly different than those presented in Table II because the standard errors were estimated differently in the
robustness model.
b
P-value for the test whether the site-to-site variance component is significantly different from zero. All are greater
than 0.05, indicating that none can be declared significantly different from zero.
tablished initial conditions in which the concentration
of bacteria ranged more than 100-fold, from common
(3 cells/mL⫺1) to rare (0.03 cells/mL⫺1). The
ScanRDI威 and the reference sterility test methods
were used to indicate presence/absence of growth in a
five-tube MPN procedure. Microbes were enumerated
using an MPN procedure because the MPN method
produces a quantitative measure of the concentration
of microorganisms in a sample even when microor-
ganisms are present at very low concentrations. The
microorganisms chosen for comparison were those
recommended by USP/EP/JP pharmacopeias and sup-
plemented with two additional organisms, E. coli and
P. acnes. E. coli was included because it is recom-
mended as a test organism for total viable aerobic
count (16), and P. acnes was included because it is a
slow-growing, anaerobic to aerotolerant bacterium Figure 2
that is frequently associated with contamination from
human flora. As such, the aerobic nature of the Percent of culture tubes showing the presence of
ScanRDI威 assay method imposes a severe metabolic microorganisms at three dilution levels. Open sym-
stress on this organism. It is clear that the limit of bols depict data collected using the ScanRDI威
detection for P. acnes was not adversely affected by method, while closed symbols depict data collected
the additional metabolic stress. We thus employed a using the reference compendial sterility test
range in the types of organisms including anaerobes method. Values alongside the names of organisms
and aerobes, Gram-negatives and Gram-positives, in the legend indicate the total number of tubes
those with fermentative metabolism and those with tested for a given organism. Horizontal lines
respiratory metabolism, and prokaryotes and eu- (dashed or dotted) depict the overall mean for all
karyotes. organisms at a given dilution.
Vol. 64, No. 4, July–August 2010 361
Downloaded from journal.pda.org on September 1, 2010
TABLE IV References
Comparison of the Limits of Detection
(cells/mLⴚ1) 1. Moldenhauer, J.; Sutton, S. W. Towards an im-
proved sterility test. PDA J. Pharm. Sci. Technol.
ScanRDI威 Compendial
2004, 58 (6), 284 –286.
Organism Method Method
Clostridium sporogenes 0.000070 0.002992 2. Halvorsen, H. O.; Ziegler, N. R. Application of
Propionibacterium acnes 0.000153 0.002805 statistics in bacteriology, I. A means of determin-
Escherichia coli 0.000805 0.003846 ing bacterial populations by the dilution method.
Pseudomonas aeruginosa 0.001172 0.017179 J. Bacteriol. 1932, 25, 101–121.
Staphylococcus aureus 0.000489 0.002871
Bacillus subtilis 0.000192 0.001832 3. Sun, X.; Kurosu, S.; Shintani, H. The expanded
Aspergillus niger 0.000690 0.003199 application of the most probable number to the
Candida albicans 0.000178 0.005370 quantitative evaluation of extremely low micro-
bial count. PDA J. Pharm. Sci. Technol. 2006, 60
(2), 124 –134.
In a recent commentary, Moldenhauer and Sutton (1, 4. Kepner Jr., R. L.; Pratt. J. R. The use of fluoro-
17) outlined some of the flaws associated with the chromes for the direct enumeration of total bac-
standard sterility test. They point out that it is well teria in environmental samples: past and present.
within the limits of current technology to conduct Microbiol. Rev. 1994, 58 (4), 603– 615.
sterility testing without the necessity of culturing or-
ganisms and the attendant problems of culturing tech- 5. Baudart, J.; Coallier, J.; Laurent, P.; Prévost,
niques (medium type, incubation conditions, detection M. Rapid and sensitive enumeration of viable
limits, etc.). The ScanRDI威 effectively avoids many of diluted cells of members of the family Enter-
the problems associated with detection of microbes by obacteriaceae in freshwater and drinking water.
culture approaches. It links two metrics of cell viabil- Appl. Environ. Microbiol. 2002, 68 (10), 5057–
ity—the action of nonspecific esterases and the pres- 5063.
ence of an intact-functioning cell-membrane—with
the production of a fluorochrome. Cells containing the 6. Jones, D.; Brailsford, M. A.; Drocourt, J.-L.
fluorochrome are easily detected by laser and subse- Solid-phase laser scanning cytometry: a new two-
quently detected (and enumerated) by computer. The hour method for the enumeration of microorgan-
approach is rapid and sensitive and allows for the isms in pharmaceutical water. Pharmacopoeial
detection of a single cell on the surface of a 25-mm Forum 1999, 25 (1), 7626 –7645.
membrane (5).
7. Onadipe, A.; Ulvedal; K. A method for the rapid
When routinely used, the ScanRDI威 requires neither detection of microbial contaminants in animal cell
lengthy incubation times nor specific culture media. culture processes. PDA J. Pharm. Sci. Technol.
However, by combining requirements of the reference 2001, 55 (6), 337–345.
sterility test (specific organisms, growth in specific
media, incubation conditions, and duration) with an 8. Ramond, B.; Rolland, X.; Planchez, C.; Cornet,
MPN enumeration protocol we were able to directly P.; Antoni, C.; Drocourt J.-L. Enumeration of
compare the two approaches to sterility testing. The total viable microorganisms in antibiotic raw ma-
ScanRDI威 method for detection of microbes was dem- terial using ChemScan威 solid phase cytometer.
onstrated to be statistically non-inferior to the refer- PDA J. Pharm. Sci. Technol. 2000, 54 (4), 320 –
ence sterility test and numerically superior in that it 331.
had a likelihood of detecting microbes that was sig-
nificantly greater at all dilution levels. As such, the 9. USP 71 Sterility Tests. In United States Pharma-
ScanRDI威 method is appropriate for use as a rapid copeia, 31st Rev.; U.S. Pharmacopeial Conven-
alternative to the growth-based sterility test method. tion, Inc.: Rockville, MD, 2008.
362 PDA Journal of Pharmaceutical Science and Technology
Downloaded from journal.pda.org on September 1, 2010
10. EP 2.6.1 Sterility. In European Pharmacopoeia, 6th 14. Jennison, C.; Turnbull, B. W. Group Sequential
ed.; European Department for the Quality of Medi- Methods with Applications to Clinical Trials;
cines and Healthcare: Strasbourg, France, 2008. Chapman and Hall/CRC: New York, 2000;
p 416.
11. Sterility Test. In Japanese Pharmacopoeia, 15th
ed.; Society of Japanese Pharmacopoeia: Tokyo, 15. SAS/STAT software, SAS System for Windows
Japan, 2006; p 93. version 9.1.3.; SAS Institute, Inc.: Cary, NC,
2001–2002.
12. Blodgett, R. Most Probable Number Determina-
tion from Serial Dilutions. In Bacteriological An-
16. EP 2.6.12 Microbial Examination of Nonsterile
alytical Manual; Jackson, G. J., Merker, R. I.,
Products: Total Viable Aerobic Count. In Euro-
Bandler, R.; BAM Project Coordinators. U.S.
pean Pharmacopoeia, 6th ed.; European Depart-
Food and Drug Administration. http://www.
fda.gov/Food/ScienceResearch/LaboratoryMethods/ ment for the Quality of Medicines and Healthcare:
BacteriologicalAnalyticalManualBAM/default.htm. Strasbourg, France, 2008.
13. USP 1223 Validation of Alternative Microbiolog- 17. Moldenhauer, J. Viability-based rapid microbiolog-
ical Methods. In United States Pharmacopeia, ical methods for sterility testing and the need for
31st Rev.; U.S. Pharmacopeial Convention, Inc.: identification of contamination. PDA J. Pharm. Sci.
Rockville, MD, 2008. Technol. 2006, 60 (2), 81– 88.
Vol. 64, No. 4, July–August 2010 363
Downloaded from journal.pda.org on September 1, 2010
An Authorized User of the electronic PDA Journal of Pharmaceutical Science and
Technology (the PDA Journal) is a PDA Member in good standing. Authorized Users are
permitted to do the following:
·Search and view the content of the PDA Journal
·Download a single article for the individual use of an Authorized User
·Assemble and distribute links that point to the PDA Journal
·Print individual articles from the PDA Journal for the individual use of an Authorized User
·Make a reasonable number of photocopies of a printed article for the individual use of an
Authorized User or for the use by or distribution to other Authorized Users
Authorized Users are not permitted to do the following:
·Except as mentioned above, allow anyone other than an Authorized User to use or access the
PDA Journal
· Display or otherwise make any information from the PDA Journal available to anyone other
than an Authorized User
·Post articles from the PDA Journal on Web sites, either available on the Internet or an Intranet,
or in any form of online publications
·Transmit electronically, via e-mail or any other file transfer protocols, any portion of the PDA
Journal
·Create a searchable archive of any portion of the PDA Journal
·Use robots or intelligent agents to access, search and/or systematically download any portion
of the PDA Journal
·Sell, re-sell, rent, lease, license, sublicense, assign or otherwise transfer the use of the PDA
Journal or its content
·Use or copy the PDA Journal for document delivery, fee-for-service use, or bulk reproduction or
distribution of materials in any form, or any substantially similar commercial purpose
·Alter, modify, repackage or adapt any portion of the PDA Journal
·Make any edits or derivative works with respect to any portion of the PDA Journal including any
text or graphics
·Delete or remove in any form or format, including on a printed article or photocopy, any
copyright information or notice contained in the PDA Journal
You might also like
- SOP For Clinical PharmacyDocument64 pagesSOP For Clinical PharmacyRaymond Ofori100% (3)
- Biomerieux ScanRDI Manual - EnglishDocument91 pagesBiomerieux ScanRDI Manual - Englishblack betty100% (1)
- Rapid Microbiology Method by Jeane MoldenhauerDocument24 pagesRapid Microbiology Method by Jeane MoldenhauerMaurits TobingNo ratings yet
- Upsteam ProcessingDocument21 pagesUpsteam ProcessingAdithyaNo ratings yet
- Sutton The Sterility TestsDocument26 pagesSutton The Sterility TestsSilke IgemannNo ratings yet
- Rapid Microbiological Methods They Are Rapid Are They FastDocument9 pagesRapid Microbiological Methods They Are Rapid Are They FastRonald SalasNo ratings yet
- Methods of Rapid Microbiological AssayDocument10 pagesMethods of Rapid Microbiological AssayMundi OdiumNo ratings yet
- En Do Toxin CatalogDocument17 pagesEn Do Toxin CatalogmarronyyyNo ratings yet
- Biologic License ApplicationDocument16 pagesBiologic License ApplicationJean Sandra PintoNo ratings yet
- The Future of Monoclonal AntibodyDocument180 pagesThe Future of Monoclonal AntibodyJohn John100% (1)
- ANDA's Impurities in Drug SubstancesDocument19 pagesANDA's Impurities in Drug SubstancesparagnkpatelNo ratings yet
- Use and Limitations of in Vitro Dissolution Testing: Topic Introduction and OverviewDocument114 pagesUse and Limitations of in Vitro Dissolution Testing: Topic Introduction and OverviewMuthu Venkatesh100% (1)
- CMCDocument3 pagesCMCSri harsha reddyNo ratings yet
- Microbiological Pharmaceutical Quality Control Labs (7 - 93) - FDADocument8 pagesMicrobiological Pharmaceutical Quality Control Labs (7 - 93) - FDAArmando SaldañaNo ratings yet
- SOP For Antimicrobial Effectiveness TestingDocument4 pagesSOP For Antimicrobial Effectiveness TestingGencay ErginNo ratings yet
- Pda: A Global Association: Recombinant Factor C - EndolisaDocument29 pagesPda: A Global Association: Recombinant Factor C - EndolisaPiruzi MaghlakelidzeNo ratings yet
- 5 1 10 Guidelines For Using The Test For Bacterial EndotoxinsDocument5 pages5 1 10 Guidelines For Using The Test For Bacterial Endotoxinsnguyentuanson167100% (1)
- FDA Guidance For Industry S7a Safety Pharmacology Studies For Human Pharmaceuticals PDFDocument14 pagesFDA Guidance For Industry S7a Safety Pharmacology Studies For Human Pharmaceuticals PDFbmartindoyle6396No ratings yet
- 1.1 Analytical Chemistry: Introductio NDocument56 pages1.1 Analytical Chemistry: Introductio NmadhubaddapuriNo ratings yet
- FDA Guidance For CMC For Clinical TrialsDocument27 pagesFDA Guidance For CMC For Clinical TrialsNelson Alejandro FierroNo ratings yet
- Ophthalmic Drug Delivery Systems, 2ed. 2003Document727 pagesOphthalmic Drug Delivery Systems, 2ed. 2003VuqarNo ratings yet
- Stability of Pharmaceuticals-1Document96 pagesStability of Pharmaceuticals-1Hely PatelNo ratings yet
- ObjectionableDocument9 pagesObjectionabledmtalbhogeNo ratings yet
- Sterile Parenteral Product (Review) PDFDocument8 pagesSterile Parenteral Product (Review) PDFdianiNo ratings yet
- FDA - ICH M7 (R1) - Control of Mutagenic Impurities in Pharmaceuticals 03.2018Document131 pagesFDA - ICH M7 (R1) - Control of Mutagenic Impurities in Pharmaceuticals 03.2018Catrinescu OanaNo ratings yet
- The New Concept of Automatic Gloved Hand SanitizationDocument3 pagesThe New Concept of Automatic Gloved Hand SanitizationTim SandleNo ratings yet
- CTD CMC-Information-Human-Gene-Therapy-IND-Applications - Jan - 2020Document56 pagesCTD CMC-Information-Human-Gene-Therapy-IND-Applications - Jan - 2020Shiraz KhanNo ratings yet
- Practical Guide To Autoclave Validation: by Raymond G. Lewis, PEDocument8 pagesPractical Guide To Autoclave Validation: by Raymond G. Lewis, PEManarKhNo ratings yet
- Guidelines For Cell LinesDocument26 pagesGuidelines For Cell LinesexecNo ratings yet
- Antimicrobial EffectivenessDocument6 pagesAntimicrobial EffectivenessChristineNo ratings yet
- Anti Microbiological Assay Test or Antibiotic Assay Test of Pharmaceutical Preparation Containing Antibiotics Using 'Cylinder Plate Method'Document4 pagesAnti Microbiological Assay Test or Antibiotic Assay Test of Pharmaceutical Preparation Containing Antibiotics Using 'Cylinder Plate Method'Editor IJTSRDNo ratings yet
- Microbial Contamination-A Regulatory Perspective.Document8 pagesMicrobial Contamination-A Regulatory Perspective.Sameer PanditNo ratings yet
- Risk Assessment in The Pharmaceutical Industry ToxicologyDocument41 pagesRisk Assessment in The Pharmaceutical Industry ToxicologyJasonNo ratings yet
- Usp 601Document33 pagesUsp 601Nur AcarNo ratings yet
- Successful Sterility Test Failure Investigations-A Practical ApproachDocument5 pagesSuccessful Sterility Test Failure Investigations-A Practical ApproachMiniq ForondaNo ratings yet
- Mammalian Expression SystemsDocument16 pagesMammalian Expression SystemsfdjkafjkljdklfjalNo ratings yet
- Valiadasi Method CleainhgDocument76 pagesValiadasi Method CleainhgYulis AdrianaNo ratings yet
- Seminar (Photostability)Document12 pagesSeminar (Photostability)Mr. HIMANSHU PALIWALNo ratings yet
- Fda 1571Document2 pagesFda 1571Nitin KashyapNo ratings yet
- Sterile Ophthalmic Preparations and Cont PDFDocument7 pagesSterile Ophthalmic Preparations and Cont PDFRaedMohNo ratings yet
- 62 The Basics of Bioburden TestingDocument2 pages62 The Basics of Bioburden Testinghitham shehataNo ratings yet
- Mark Old Corne Microbiological ValidationDocument28 pagesMark Old Corne Microbiological ValidationhumusdelombrizNo ratings yet
- Cundell Et Al PDA J Pharm Sci Tech March 2010 (Full Article) SfsDocument21 pagesCundell Et Al PDA J Pharm Sci Tech March 2010 (Full Article) SfsJessica González LaraNo ratings yet
- Gamma IrradiationDocument27 pagesGamma Irradiationmuzammil21_adNo ratings yet
- 1111 - USP Micro Limit Test For Non SterileDocument2 pages1111 - USP Micro Limit Test For Non SterileSpectre Spectre100% (1)
- Patent-US 20070003628 A1-Nanoparticulate Clopidogrel FormulationsDocument19 pagesPatent-US 20070003628 A1-Nanoparticulate Clopidogrel FormulationsQuan NguyenNo ratings yet
- PUPSITDocument43 pagesPUPSITAlessandro ChreimNo ratings yet
- PQLI Definition of CriticalityDocument10 pagesPQLI Definition of CriticalitypakdekroNo ratings yet
- 23635.1.5. Application of The F0 Concept To Steam Sterilisation of Aqueou - PDFDocument1 page23635.1.5. Application of The F0 Concept To Steam Sterilisation of Aqueou - PDFlilaNo ratings yet
- Quality Control and Drug AnalysisDocument57 pagesQuality Control and Drug AnalysisIhab AdelNo ratings yet
- SFDA Saudi Biosimilar GuidelinesDocument17 pagesSFDA Saudi Biosimilar Guidelinesshivani hiremath100% (1)
- Preventing Metal ContaminationDocument6 pagesPreventing Metal Contaminationlouish9175841No ratings yet
- Development of New Ophthalmic Suspension Prednisolone AcetateDocument6 pagesDevelopment of New Ophthalmic Suspension Prednisolone AcetaterandatagNo ratings yet
- Old Drugs For A New Use (Prescription)Document17 pagesOld Drugs For A New Use (Prescription)Karol Denisse Hernández BastidaNo ratings yet
- Selection of DissolutionDocument5 pagesSelection of DissolutionGirishNo ratings yet
- USP Seminar - Fundamentals of Bioassay Practices 2014 PDFDocument140 pagesUSP Seminar - Fundamentals of Bioassay Practices 2014 PDFnsk79in@gmail.comNo ratings yet
- Us P 797 Pharmaceutical Compounding Sterile CompoundingDocument61 pagesUs P 797 Pharmaceutical Compounding Sterile Compoundingdeepa_ragu5695No ratings yet
- Burkholderia CepaciaDocument11 pagesBurkholderia Cepaciaportner6873No ratings yet
- Container Closure IntegrityDocument19 pagesContainer Closure IntegrityYessieNo ratings yet
- A Comprehensive Text Book on Self-emulsifying Drug Delivery SystemsFrom EverandA Comprehensive Text Book on Self-emulsifying Drug Delivery SystemsNo ratings yet
- Enzyme Inhibition in Drug Discovery and Development: The Good and the BadFrom EverandEnzyme Inhibition in Drug Discovery and Development: The Good and the BadChuang LuNo ratings yet
- Exercise Cost of CapitalDocument7 pagesExercise Cost of CapitalUmair Shekhani100% (2)
- Exercise Capital BudgetingDocument2 pagesExercise Capital BudgetingUmair ShekhaniNo ratings yet
- Exercise Capital BudgetingDocument2 pagesExercise Capital BudgetingUmair ShekhaniNo ratings yet
- Balanced ScorecardDocument56 pagesBalanced ScorecardUmair ShekhaniNo ratings yet
- RD of CBD ProductsDocument13 pagesRD of CBD ProductsUmair ShekhaniNo ratings yet
- Assignment # 2: Book: From Financial Accounting, Ninth Edition, by Weygandt, Kimmel, KiesoDocument1 pageAssignment # 2: Book: From Financial Accounting, Ninth Edition, by Weygandt, Kimmel, KiesoUmair ShekhaniNo ratings yet
- Business CommunicationDocument18 pagesBusiness CommunicationUmair ShekhaniNo ratings yet
- Exercise Stock ValuationDocument2 pagesExercise Stock ValuationUmair ShekhaniNo ratings yet
- Commercial Facility For CBD ProductsDocument6 pagesCommercial Facility For CBD ProductsUmair ShekhaniNo ratings yet
- A. Introduction / Background: Industrial and Medicinal Management of Hemp As An Agriculture Commodity by Indus GroupDocument4 pagesA. Introduction / Background: Industrial and Medicinal Management of Hemp As An Agriculture Commodity by Indus GroupUmair ShekhaniNo ratings yet
- Adr Form 1Document2 pagesAdr Form 1Umair ShekhaniNo ratings yet
- Adr Form 2Document2 pagesAdr Form 2Umair ShekhaniNo ratings yet
- Indus Pharma (PVT.) LTD.: Protocol For Re-Qualification of Water Distillation Plant (Wfi), Storage & Distribution SystemDocument9 pagesIndus Pharma (PVT.) LTD.: Protocol For Re-Qualification of Water Distillation Plant (Wfi), Storage & Distribution SystemUmair Shekhani100% (1)
- Inspection Report of Incoming PMDocument1 pageInspection Report of Incoming PMUmair ShekhaniNo ratings yet
- Standard Operating Procedure: Indus Pharma (PVT.) LTDDocument4 pagesStandard Operating Procedure: Indus Pharma (PVT.) LTDUmair ShekhaniNo ratings yet
- Temperature Mapping DataDocument13 pagesTemperature Mapping DataUmair ShekhaniNo ratings yet
- Standard Operating Procedure: Indus Pharma (PVT.) LTDDocument4 pagesStandard Operating Procedure: Indus Pharma (PVT.) LTDUmair ShekhaniNo ratings yet
- Indus Pharma (PVT.) LTD.: Complaint Response FormDocument1 pageIndus Pharma (PVT.) LTD.: Complaint Response FormUmair ShekhaniNo ratings yet
- EMA Epar Public Assessment ReportDocument37 pagesEMA Epar Public Assessment ReportOscar Javier EstupiñánNo ratings yet
- Formula Ambroxol JurnalDocument3 pagesFormula Ambroxol Jurnalrd_al_snrNo ratings yet
- Summer Training Report AbhiDocument51 pagesSummer Training Report AbhiAbhijit MohantyNo ratings yet
- Mimusops Elengi Linn:a ReviewDocument9 pagesMimusops Elengi Linn:a ReviewRAPPORTS DE PHARMACIENo ratings yet
- Usp 36-NF 31 - GC41Document1 pageUsp 36-NF 31 - GC41DennyNo ratings yet
- Why Companies Are Selling Same Drug in Different PricesDocument2 pagesWhy Companies Are Selling Same Drug in Different Pricesdharmendra gaurNo ratings yet
- Pharmacist Syllabus DetailedDocument18 pagesPharmacist Syllabus DetailedHari Kamesh KiranNo ratings yet
- HhihhygtfrtdgbhjDocument11 pagesHhihhygtfrtdgbhjKania HidayantiNo ratings yet
- GCC Data Requirements For Human Drugs Submission Version 1.1 PDFDocument81 pagesGCC Data Requirements For Human Drugs Submission Version 1.1 PDFBasha Yazn Anjak50% (2)
- uk-spc-nexium-sachet-10mg-DRESS-update-GI 21 0012aDocument17 pagesuk-spc-nexium-sachet-10mg-DRESS-update-GI 21 0012aDeisy ClerkeNo ratings yet
- Jurnal LUTS PDFDocument8 pagesJurnal LUTS PDFmitaNo ratings yet
- Deflazen: Training ManualDocument10 pagesDeflazen: Training Manualanupdr_cNo ratings yet
- Medco Express FINALDocument23 pagesMedco Express FINALSean MooreNo ratings yet
- Three Different Types of Airlocks in Pharmaceuticals - Pharma PathwayDocument4 pagesThree Different Types of Airlocks in Pharmaceuticals - Pharma PathwayFaizul ZainudinNo ratings yet
- ExperimentDocument9 pagesExperimentsatyakrishna1234No ratings yet
- List of Cos in AhmedabadDocument559 pagesList of Cos in AhmedabadVikhyat SharmaNo ratings yet
- Drug Development Process - Part 1Document5 pagesDrug Development Process - Part 1Nguyễn Thế ThaoNo ratings yet
- Pharm MCQDocument17 pagesPharm MCQArun Kumar100% (2)
- Cough Syrup Detail:: Prodactive or Expectorant and Suppressant or Dry CoughDocument3 pagesCough Syrup Detail:: Prodactive or Expectorant and Suppressant or Dry CoughFarhadullah KhanNo ratings yet
- Barros 2014 - The Use of Pictograms in The Health CareDocument16 pagesBarros 2014 - The Use of Pictograms in The Health CareBlaberNo ratings yet
- Drug Delivery SystemsDocument8 pagesDrug Delivery SystemsNikko Nabasca GorneNo ratings yet
- List of Abbreviations Used in Medical PrescriptionsDocument5 pagesList of Abbreviations Used in Medical PrescriptionsNathan TonthatNo ratings yet
- Lista de Precios Farmacos 27-04-2022Document5 pagesLista de Precios Farmacos 27-04-2022Mery Carolina PerezNo ratings yet
- The Science of Ischemic Stroke Pathophysiology Pharmacological TreatmentDocument20 pagesThe Science of Ischemic Stroke Pathophysiology Pharmacological TreatmentLisa Linggi'AlloNo ratings yet
- Switching P2Y Inhibitors: Medication SafetyDocument1 pageSwitching P2Y Inhibitors: Medication SafetyWaleed saleemNo ratings yet
- Presented By: Samiksha B.Sawant M, PHARM (IP), 1 SEM Guided By: Dr. Indira ParabDocument46 pagesPresented By: Samiksha B.Sawant M, PHARM (IP), 1 SEM Guided By: Dr. Indira ParabKainat MalikNo ratings yet
- Pharmacy Daily For Wed 09 Sep 2015 - Australia-China Drug Pact, Pharmacy Continence Courses, TWC Hearing Tests, Health AMPERSAND Beauty and Much MoreDocument4 pagesPharmacy Daily For Wed 09 Sep 2015 - Australia-China Drug Pact, Pharmacy Continence Courses, TWC Hearing Tests, Health AMPERSAND Beauty and Much MorepharmacydailyNo ratings yet
- Friends PharmaDocument3 pagesFriends PharmaaoryaNo ratings yet
- 70-Article Text-286-2-10-20221205Document7 pages70-Article Text-286-2-10-20221205Nurul Maulida PutriNo ratings yet