Professional Documents
Culture Documents
Module 4 - Enzymes 1 PDF
Module 4 - Enzymes 1 PDF
Uploaded by
Francis ValdezOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Module 4 - Enzymes 1 PDF
Module 4 - Enzymes 1 PDF
Uploaded by
Francis ValdezCopyright:
Available Formats
MODULE 5 ENZYMES
Module 4
Module
4
MTCC 117
Enzymes 1
Charisse Anne P. Igaran, RMT
JMJ Marists Brothers
Notre Dame of Marbel University
College of Arts and Sciences
MTCC117/INSTRUCTOR:
MEDICALCHARISSE
TECHNOLOGYANNE PASION IGARAN, RMT
DEPARTMENT 1ST SEMESTER S.Y.2020-2021 1
MODULE 5 ENZYMES
Virtually all reactions in the body are mediated by enzymes, which are protein catalysts that increase
the rate of reactions without being changed in the overall process. Among the many biologic reactions that
are energetically possible, enzymes selectively channel reactants (called substrates) into useful pathways.
Enzymes thus direct all metabolic events. This module examines the nature of these catalytic molecules and
their mechanism of action.
LEARNING OBJECTIVES
At the end of the module, the students will be able to:
1. Discuss general characteristics for enzymes in terms of structure and
composition
2. Describe what a substrate is and be familiar with the terminology used to
classify enzymes based on the types of reactions they catalyse.
3. Describe the concept of a “lock and key” and “induced fit” model of
enzyme action.
4. Be able to categorize enzymes in terms of specificity characteristics.
5. Explain how temperature, pH, substrate concentration, and enzyme
concentration affect enzyme activity.
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 2
MODULE 5 ENZYMES
ABSTRACTION
General Characteristics of Enzymes
• Enzymes are usually proteins that act as biological catalysts.
• Each cell in the human body contains thousands of different enzymes.
• Enzymes cause cellular reactions to occur millions of times faster than corresponding uncatalyzed reactions
• An enzyme speeds a reaction by lowering the activation energy, changing the reaction pathway that provides a
lower energy route for conversion of substrate to product.
• As catalysts enzymes are not consumed in the reactions
• A few enzymes are now known to be ribonucleic acids (RNA)
Enzyme Structure
Simple and Conjugated Enzymes
• Most enzymes are globular proteins; some are simple proteins, others are conjugated proteins
• Simple enzyme: composed only of protein (amino acid chains)
It is the 3o structure of the simple enzymes that makes it biologically active
• Conjugated enzyme: has a non-protein part in addition to a protein part.
1. apoenzyme -protein part; inactive in itself
2. cofactor /coenzyme- nonprotein organic
(coenzyme /co-substrate) or inorganic (cofactor) moiety; the activator; loosely bound to protein
•apoenzyme + cofactor = holoenzyme (biologically active conjugated enzyme)
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 3
MODULE 5 ENZYMES
Nomenclature and Classification of Enzymes
• Most commonly named with reference to their function
– type of reaction catalyzed
– identity of the substrate
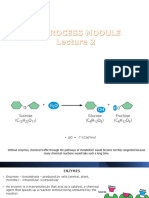
• A substrate is the reactant in an enzyme-catalyzed reaction:
– the substrate is the substance upon which the enzyme “acts.”
– e. g., In the fermentation process, sugar is converted to alcohol, therefore in this reaction sugar is the
substrate
Three Important Aspects of the Naming
Process
1. Suffix -ase identifies it as an enzyme
a. e.g., urease, sucrase, and lipase are
all enzyme designations
b. exception: the suffix -in is still
found in the names of some
digestive enzymes, e.g., trypsin,
chymotrypsin, and pepsin
2. Type of reaction catalyzed by an
enzyme is often used as a prefix
c. e.g., oxidase - catalyzes an
oxidation reaction,
d. e.g., hydrolase - catalyzes a hydrolysis reaction
3. Identity of substrate is often used in addition to the type of reaction
e. e.g. glucose oxidase, pyruvate carboxylase, and succinate dehydrogenase
PRACTICE EXERCISE
1. Predict the function of the following enzymes.
a. Maltase
b. Lactate dehydrogenase
c. Fructose oxidase
d. Maleate isomerase
2. Predict the function of the following enzymes.
a. Maltase
b. Lactate dehydrogenase
c. Fructose oxidase
d. Maleate isomerase
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 4
MODULE 5 ENZYMES
Six Major Classes of Enzymes
• Enzymes are grouped into six major classes based on the types of reactions they catalyse
Class Reaction Catalyzed
1. Oxidoreductases Oxidation-reductions
To help you
2. Transferases Functional group transfer reactions
remember,
3. Hydrolases Hydrolysis reactions
4. Lyases Reactions involving addition of a group
“O-T-H-L-I-L”
to a double bond or removal of groups to
form double bonds
5. Isomerase Isomerization reactions
6. Ligases Reactions involving bond formation
coupled with ATP
hydrolysis
Oxidoreductase
• An oxidoreductase enzyme catalyzes an oxidation–reduction reaction: Up one, down one
– oxidation and reduction reactions are always linked to one another
– an oxidoreductase requires a coenzyme that is either oxidized or reduced as the substrate in the
reaction.
– e.g., lactate dehydrogenase is an oxidoreductase and NAD+ is the coenzyme in this reaction.
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 5
MODULE 5 ENZYMES
Transferase
• A transferase is an enzyme that catalyzes the transfer of a functional group You don’t belong
from one molecule to another here!
• Two major subtypes:
1. kinases - catalyze transfer of a phosphate group from adenosine triphosphate (ATP) to a substrate
2. transaminases - catalyze transfer of an amino group to a substrate
Hydrolase
Water does it again!
• a hydrolase is an enzyme that catalyzes a hydrolysis reaction
• the reaction involves addition of a water molecule to a bond to cause bond breakage
• hydrolysis reactions are central to the process of digestion:
– carbohydrases hydrolyze glycosidic bonds in oligo- and polysaccharides
– proteases effect the breaking of peptide linkages in proteins
– lipases effect the breaking of ester linkages in triacylglycerols
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 6
MODULE 5 ENZYMES
Lyase Taking it apart
• A lyase is an enzyme that catalyzes the addition or the removal of a group
in a manner that does not involve hydrolysis or oxidation
– dehydratase: effects the removal of the components of water to form a double bond
– hydratase: effects the addition of the components of water to a double bond
– decarboxylase: effects the removal of carbon dioxide from a substrate
– deaminase: effects the removal of ammonia from a substrate
Isomerase
• An isomerase is an enzyme that catalyzes the isomerization (rearrangement
of atoms) of a substrate in a reaction, converting it into a molecule isomeric
Shuffling the deck
with itself.
– racemases – conversion of D- to L- isomer or vice versa
– mutases – transfer of a functional group within a molecule
Ligase
• A ligase is an enzyme that catalyzes the formation of a bond between two Putting it together
molecules involving ATP hydrolysis to ADP:
– ATP hydrolysis is required because such reactions are energetically unfavorable
– synthetases – formation of new bond between two substrates with participation of ATP
– carboxylases – formation of new bond between substrate and carbon dioxide with participation of ATP
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 7
MODULE 5 ENZYMES
PRACTICE EXERCISE
To what main enzyme class do the enzymes that catalyze the
following chemical reactions belong?
From the keywords we used to remember the class of
enzymes, try to remember the enzymes with these phrases.
Up one, down one You don’t belong Water does it again!
here!
Putting it together Taking it apart Shuffling the deck
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 8
MODULE 5 ENZYMES
Models of Enzyme Action
• Enzyme Active Site
• Explanations of how enzymes function as catalysts in
biochemical systems are based on the concepts of an enzyme
active site and enzyme-substrate complex formation.
• The active site: relatively small part of an enzyme’s structure
that is actually involved in catalysis:
– where substrate binds to enzyme
– formed due to folding and bending of the protein.
– usually a “crevice like” location in the enzyme
– some enzymes have more than one active site
Enzyme Substrate Complex
• Intermediate reaction species formed when substrate binds
with the active site
• Needed for the activity of enzyme
• Orientation and proximity is favorable and reaction is
fast
ACTIVITY 1
To help you visualize the enzyme-substrate complex, please copy the link and watch the video,
https://www.youtube.com/watch?v=aRSfPLp_I10
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 9
MODULE 5 ENZYMES
Two Models for Substrate Binding to Enzyme
• Lock-and-Key model:
– In this model, the active site in the enzyme has a fixed, rigid geometrical conformation
– only substrate of specific shape can bind with active site; a substrate whose shape and chemical nature
are complementary to those of the active site can interact with the enzyme.
– fails to take into account proteins’ conformational changes to accommodate a substrate molecule
• Induced Fit Model:
– substrate contact with enzyme will change the shape of the active site
– allows small change in space to accommodate substrate (e.g., how a hand fits into a glove)
– the enzyme active site, although not exactly complementary in shape to that of the substrate, is flexible
enough that it can adapt to the shape of the substrate.
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021
10
MODULE 5 ENZYMES
Forces That Determine Substrate Binding
• H-bonding
• Hydrophobic
interactions
• Electrostatic
interactions
Enzyme Specificity
• Absolute Specificity:
– an enzyme will catalyze a particular reaction for only one substrate
– this is most restrictive of all specificities (not common)
– e.g., catalase is an enzyme with absolute specificity for hydrogen peroxide (H2O2)
– urease absolute specificity for urea
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021 11
MODULE 5 ENZYMES
• Stereochemical Specificity:
– an enzyme can distinguish between stereoisomers
– chirality is inherent in an active site (amino acids are chiral compounds)
– L-amino-acid oxidase - catalyzes reactions of L-amino acids but not of D-amino acids.
• Group Specificity:
– involves structurally similar compounds that have the same functional groups.
– e.g., carboxypeptidase: cleaves amino acids one at a time from the carboxyl end of the peptide chain
• Linkage Specificity:
– involves a particular type of bond irrespective of the structural features in the vicinity of the bond
– considered most general of enzyme specificities
– e.g., phosphatases: hydrolyze phosphate–ester bonds in all types of phosphate esters
Factors That Affect Enzyme Activity
Enzymes can be isolated from cells and their properties studied in a test tube (that is, in vitro). Different enzymes
show different responses to changes in substrate concentration, temperature, and pH. This section describes factors
that influence the reaction velocity of enzymes. Enzymic responses to these factors give us valuable clues as to how
enzymes function in living cells (that is, in vivo).
Enzyme Activity
• A measure of the rate at which enzyme converts
substrate to products in a biochemical reaction
• Four factors affect enzyme activity:
o Temperature
o pH
o Substrate concentration
o Enzyme concentration
Temperature
• Higher temperature results in higher kinetic energy which
causes an increase in
number of reactant collisions, therefore there is higher
activity.
• Optimum temperature: temperature at which the rate of
enzyme- catalyzed
reaction is maximum
• Optimum temperature for human enzymes is 37ºC (body
temperature)
• Increased temperature (high fever) leads to decreased
enzyme activity
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021
12
MODULE 5 ENZYMES
pH
• Drastic changes in pH can result in denaturation of proteins
• Optimum pH: pH at which enzyme has maximum activity
• Most enzymes have optimal activity in the pH range of 7.0 - 7.5
• Exception: digestive enzymes
– pepsin: optimum pH = 2.0
– trypsin: optimum pH = 8.0
Substrate Concentration
• At a constant enzyme concentration, the enzyme
activity increases with
increased substrate concentration.
• Enzyme saturation: the concentration at which it
reaches its maximum
rate and all of the active sites are full
• Turnover number: number of substrate molecules
converted to product
per second per enzyme molecule under
conditions of optimum temperature and pH
Enzyme Concentration
• Enzymes are not consumed in the reactions they
catalyze
• At a constant substrate concentration, enzyme
activity increases with
• increase in enzyme concentration
– the greater the enzyme concentration, the
greater the reaction rate.
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021
13
MODULE 5 ENZYMES
Practice Exercise
• Describe the effect that each of the following changes would have on the rate
of a reaction that involves the substrate sucrose and the intestinal enzyme sucrase.
o Decreasing the sucrase concentration
o Increasing the sucrose concentration
o Lowering the temperature to 10ºC
o Raising the pH from 6.0 to 8.0 when the optimum pH is 6.2
SUPPLEMENTARY ACTIVITY
Copy the link and watch this video and appreciate how enzymes work!
https://www.youtube.com/watch?v=yk14dOOvwMk
MTCC117/INSTRUCTOR: CHARISSE ANNE PASION IGARAN, RMT 1ST SEMESTER S.Y.2020-2021
14
You might also like
- Biochemical Energetics: Biochemistry of MetabolismDocument36 pagesBiochemical Energetics: Biochemistry of MetabolismDozdiNo ratings yet
- 1 - FST-607. Food Biotechnology (L # 1-10) - RepublishedDocument112 pages1 - FST-607. Food Biotechnology (L # 1-10) - RepublishedZubair JuttNo ratings yet
- SAP ATP CalculationDocument6 pagesSAP ATP Calculationaprian100% (1)
- Enzymes CoenzymesDocument71 pagesEnzymes CoenzymesAiswarya ManojkumarNo ratings yet
- Unit 7 EnzymesDocument85 pagesUnit 7 EnzymesAngelica Camille B. AbaoNo ratings yet
- 1-Introduction To EnzymesDocument25 pages1-Introduction To EnzymesMarwa Mohamed AminNo ratings yet
- EnzymeDocument192 pagesEnzymeKishoreNo ratings yet
- EnzymesDocument46 pagesEnzymesHighlifeNo ratings yet
- Chapter 4 Enzymes and VitaminsDocument9 pagesChapter 4 Enzymes and Vitaminsvictoria cablayNo ratings yet
- Enzymes Part 1Document17 pagesEnzymes Part 1drdaisy2003No ratings yet
- Enzymes - BiochemistryDocument40 pagesEnzymes - Biochemistrysunil patelNo ratings yet
- EnzymeDocument15 pagesEnzymeSheena Mhae LopeñaNo ratings yet
- 3.5 Enzymes 2Document64 pages3.5 Enzymes 2Alondra SagarioNo ratings yet
- Stocker Enzymes and VitaminsDocument75 pagesStocker Enzymes and VitaminsDylan SalesNo ratings yet
- Biochemistry NCMA113 Midterm Notes PDocument3 pagesBiochemistry NCMA113 Midterm Notes PVaishnavi LoganathanNo ratings yet
- Enzymes and Vitamins NotesDocument14 pagesEnzymes and Vitamins NotesRealyn ZambasNo ratings yet
- 5enzymes and Vitamins PDFDocument48 pages5enzymes and Vitamins PDFRomelyn AngelNo ratings yet
- Nomenclature and Classification of Enzyme: Shaina Mae P. MapulaDocument17 pagesNomenclature and Classification of Enzyme: Shaina Mae P. MapulaCindy MariscotesNo ratings yet
- S5 Bio (Enzymes)Document16 pagesS5 Bio (Enzymes)juniormugarura5No ratings yet
- Enzymes PPTDocument40 pagesEnzymes PPTJaisy PatelNo ratings yet
- EnzymeDocument54 pagesEnzymeSATVIK 11No ratings yet
- Reporter 3 - EnzymesDocument77 pagesReporter 3 - EnzymesJowe VarnalNo ratings yet
- Chem113 LecDocument15 pagesChem113 LecLindon TibleNo ratings yet
- Biochemistry OralsDocument214 pagesBiochemistry Oralsdr.mumtaz09No ratings yet
- Roles of Enzymes in Metabolic ReactionsDocument42 pagesRoles of Enzymes in Metabolic ReactionsAteca TabualevuNo ratings yet
- L-1 EnzymesDocument3 pagesL-1 EnzymesLiyakath AliNo ratings yet
- Chapter 12 EnymesDocument6 pagesChapter 12 EnymesveymaramaNo ratings yet
- Lecture 2Document18 pagesLecture 2warden tambiNo ratings yet
- BT1301 NolDocument131 pagesBT1301 NolJanani RipplingrythmNo ratings yet
- EnzymesDocument58 pagesEnzymesFateh RaufNo ratings yet
- Enzymology To Kreb's CycleDocument34 pagesEnzymology To Kreb's Cycleferdinand padillaNo ratings yet
- Enzymes For BPT StudentsDocument68 pagesEnzymes For BPT Studentsilagernischit321No ratings yet
- Week 9 - EnzymesDocument5 pagesWeek 9 - Enzymesjvlegaspi7463valNo ratings yet
- Enzymes PPTDocument39 pagesEnzymes PPTsunil patelNo ratings yet
- EnzymologyDocument13 pagesEnzymologyRane MandapatNo ratings yet
- Enzymes: Enzymes Homeostasis Activation EnergyDocument6 pagesEnzymes: Enzymes Homeostasis Activation EnergyGowthami MarreddyNo ratings yet
- 1FFF11B1BD16627EE05400144FEB5F70.pptDocument63 pages1FFF11B1BD16627EE05400144FEB5F70.pptNur AishaNo ratings yet
- Biology Useful 3 Grade 11 Reference GuideDocument77 pagesBiology Useful 3 Grade 11 Reference GuideHassenMohNo ratings yet
- EnzymesDocument40 pagesEnzymesAmal100% (2)
- Enzymes in Food IndustryDocument67 pagesEnzymes in Food Industryamrutha tkNo ratings yet
- UNIT 5 WorksheetDocument10 pagesUNIT 5 WorksheetlorNo ratings yet
- Động Hoá Học Duoc-32-112Document81 pagesĐộng Hoá Học Duoc-32-112Nguyễn Đình TrườngNo ratings yet
- Enzymes LectDocument117 pagesEnzymes LectGadar NishantNo ratings yet
- Enzyme: Ayesha Shafi Pharm-D, (P.U.), M. Phil. Pharmaceutical Chemistry (P.U.)Document34 pagesEnzyme: Ayesha Shafi Pharm-D, (P.U.), M. Phil. Pharmaceutical Chemistry (P.U.)Shafaqat Ghani Shafaqat Ghani100% (2)
- Biological Molecules: Prepared By: Mrs. Eden C. SanchezDocument50 pagesBiological Molecules: Prepared By: Mrs. Eden C. SanchezBernard D. Fajardo Jr.100% (1)
- EnzymeDocument3 pagesEnzymesophiamariefoster04No ratings yet
- 222 Unit 3Document103 pages222 Unit 3Ning BalderasNo ratings yet
- Enzym HappyDocument47 pagesEnzym HappyAnggitsb NainggolanNo ratings yet
- Enzyme 1Document115 pagesEnzyme 1fikaduNo ratings yet
- Enzymes and VitaminsDocument19 pagesEnzymes and VitaminsJoanna Marie TulioNo ratings yet
- Biological Chemistry: Enzyme Kinetics Part 1Document30 pagesBiological Chemistry: Enzyme Kinetics Part 1Mohammed shaffiqueNo ratings yet
- Enzymes and Vitamins CompressedDocument14 pagesEnzymes and Vitamins CompressedfredkaseydiNo ratings yet
- OUTLINE8Document8 pagesOUTLINE8Hjm ArNo ratings yet
- 02 EnzymesDocument63 pages02 EnzymesFatish BanguraNo ratings yet
- Enzymes. RagaviDocument63 pagesEnzymes. Ragavikavi bharathiNo ratings yet
- Chapter 3 Enzymes Hormones VitaminsDocument133 pagesChapter 3 Enzymes Hormones VitaminsTran Danh NhanNo ratings yet
- Enzymes Lecture 8Document27 pagesEnzymes Lecture 8Eng.Mohamed HamedNo ratings yet
- SEMESTER AWAL 2017/2018: Dasar-Dasar Biokimia EnzimDocument38 pagesSEMESTER AWAL 2017/2018: Dasar-Dasar Biokimia EnzimFachril ismailNo ratings yet
- Biochemistry Week 3 - EnzymesDocument6 pagesBiochemistry Week 3 - EnzymesMicah JadeNo ratings yet
- Enzymes Mechanism of Enzyme ActionDocument6 pagesEnzymes Mechanism of Enzyme Actionkl42c4300No ratings yet
- Nzymes: By: Mrs. Kalaivani Sathish. M. Pharm, Assistant Professor, Pims - PanipatDocument63 pagesNzymes: By: Mrs. Kalaivani Sathish. M. Pharm, Assistant Professor, Pims - Panipaturmila pandeyNo ratings yet
- Enzymes LectureDocument120 pagesEnzymes LectureHERSEY MIAYONo ratings yet
- A-level Biology Revision: Cheeky Revision ShortcutsFrom EverandA-level Biology Revision: Cheeky Revision ShortcutsRating: 5 out of 5 stars5/5 (5)
- Updated lp2Document75 pagesUpdated lp2Francis ValdezNo ratings yet
- Antimicrobial Susceptibility Test: YeprmtmdDocument41 pagesAntimicrobial Susceptibility Test: YeprmtmdFrancis ValdezNo ratings yet
- Act.1 RQ 2 Pipettes and Others 1-3Document3 pagesAct.1 RQ 2 Pipettes and Others 1-3Francis Valdez100% (1)
- Activity No. 13: Gram Negative Pyogenic CocciDocument10 pagesActivity No. 13: Gram Negative Pyogenic CocciFrancis ValdezNo ratings yet
- Activity No. 14Document14 pagesActivity No. 14Francis ValdezNo ratings yet
- Urinec Ulture PDFDocument9 pagesUrinec Ulture PDFFrancis ValdezNo ratings yet
- Post Lab Gram Positive Pyogenic Cocci: Activity 12Document10 pagesPost Lab Gram Positive Pyogenic Cocci: Activity 12Francis ValdezNo ratings yet
- AUOBF Manual Activity 3Document6 pagesAUOBF Manual Activity 3Francis ValdezNo ratings yet
- Urinec Ulture PDFDocument9 pagesUrinec Ulture PDFFrancis ValdezNo ratings yet
- Liebermann-Burchard Test For Cholesterol: by Sherwin Carl R. DimacutacDocument13 pagesLiebermann-Burchard Test For Cholesterol: by Sherwin Carl R. DimacutacFrancis ValdezNo ratings yet
- Bacillus Clostridium SPPPDF - 2 PDFDocument68 pagesBacillus Clostridium SPPPDF - 2 PDFFrancis ValdezNo ratings yet
- The Efficacy of Mangosteen (Garcinia Mangostana) Peel Extract As An Alternative Stain To Eosin in Wright'S StainDocument15 pagesThe Efficacy of Mangosteen (Garcinia Mangostana) Peel Extract As An Alternative Stain To Eosin in Wright'S StainFrancis ValdezNo ratings yet
- Activity 12: LipidsDocument19 pagesActivity 12: LipidsFrancis ValdezNo ratings yet
- Research PPT 101Document20 pagesResearch PPT 101Francis ValdezNo ratings yet
- Review Question No. 4: By: Valerie Anne G. Driz BSMT-2CDocument12 pagesReview Question No. 4: By: Valerie Anne G. Driz BSMT-2CFrancis ValdezNo ratings yet
- Research PPT SemifinalDocument17 pagesResearch PPT SemifinalFrancis ValdezNo ratings yet
- Schedule of Defense DEC 10 and 11Document4 pagesSchedule of Defense DEC 10 and 11Francis ValdezNo ratings yet
- Streak Plate and Spread Plate Inoculation: Activity No.6Document3 pagesStreak Plate and Spread Plate Inoculation: Activity No.6Francis ValdezNo ratings yet
- The Chemical Examination of Urine: (Pick The Date) (Edition 1, Volume 1)Document11 pagesThe Chemical Examination of Urine: (Pick The Date) (Edition 1, Volume 1)Francis ValdezNo ratings yet
- Act 2 and 3 ReviewDocument7 pagesAct 2 and 3 ReviewFrancis ValdezNo ratings yet
- Activity No .5 Culture Transfer TechniquesDocument4 pagesActivity No .5 Culture Transfer TechniquesFrancis ValdezNo ratings yet
- Physical Examination of Urine: ClarityDocument4 pagesPhysical Examination of Urine: ClarityFrancis ValdezNo ratings yet
- Memo No. 8 - OfficialDocument1 pageMemo No. 8 - OfficialFrancis ValdezNo ratings yet
- Amniotic Fluid: Group 5Document29 pagesAmniotic Fluid: Group 5Francis ValdezNo ratings yet
- Fresh-Fixed Vs Air-Dried FNA Biopsy SpecimensDocument2 pagesFresh-Fixed Vs Air-Dried FNA Biopsy SpecimensFrancis ValdezNo ratings yet
- Histologic Plant Tissue Preparation Techniques. 5 - Embedding StepDocument4 pagesHistologic Plant Tissue Preparation Techniques. 5 - Embedding StepFrancis ValdezNo ratings yet
- Analytical Methods: (PART 1)Document52 pagesAnalytical Methods: (PART 1)Francis ValdezNo ratings yet
- Hemoglobin Metabolism: 1 HeyzgranDocument48 pagesHemoglobin Metabolism: 1 HeyzgranFrancis ValdezNo ratings yet
- LEC 5 RBC METABOLISM AND Membrane StructureDocument30 pagesLEC 5 RBC METABOLISM AND Membrane StructureFrancis ValdezNo ratings yet
- Excretion of Lactose in Urine As A Measure of Increased Permeability of The Lactating Breast During InflammationDocument6 pagesExcretion of Lactose in Urine As A Measure of Increased Permeability of The Lactating Breast During InflammationFrancis ValdezNo ratings yet
- Biology FAQDocument6 pagesBiology FAQAirJeans GamingNo ratings yet
- Introduction To Cell Respiration LaboratoryDocument15 pagesIntroduction To Cell Respiration LaboratoryNor safikahNo ratings yet
- SL Paper 2: Do Not Accept Energy StorageDocument73 pagesSL Paper 2: Do Not Accept Energy StoragemanrajNo ratings yet
- Enzy. Defi. Class.Document67 pagesEnzy. Defi. Class.Amrit LalNo ratings yet
- Concept 7Document3 pagesConcept 7api-533564885No ratings yet
- Cellular RespirationDocument28 pagesCellular RespirationPrecy Ann ReyesNo ratings yet
- 26th Biosciences Quiz Bee ReviewerDocument12 pages26th Biosciences Quiz Bee ReviewerEuan Joshua B. Evangelista100% (1)
- Respiration 5.2.1Document11 pagesRespiration 5.2.1bexNo ratings yet
- Unit 6 Notes Csir NetDocument163 pagesUnit 6 Notes Csir NetAlma VNo ratings yet
- Shs Pe Grade-11 1st-Semester First-QuarterDocument24 pagesShs Pe Grade-11 1st-Semester First-QuarterMarisol TapadoNo ratings yet
- Objective General KnowledgeDocument267 pagesObjective General KnowledgePriya Sridharan100% (1)
- Chapter 05Document80 pagesChapter 05John Phillip DionisioNo ratings yet
- Kumpulan Soal Bioenergetika TugasDocument19 pagesKumpulan Soal Bioenergetika TugasAnika Kunthi HutamiNo ratings yet
- A Detailed Lesson Plan in BiochemistryDocument8 pagesA Detailed Lesson Plan in BiochemistryMusa BuwatNo ratings yet
- Appolo TNPSC Vao Answer Keys 2016Document16 pagesAppolo TNPSC Vao Answer Keys 2016Dinesh KumarNo ratings yet
- In Aging Neuroscience (HTTPS://WWW - Frontie: Author Contributions Funding Con Ict of Interest ReferencesDocument33 pagesIn Aging Neuroscience (HTTPS://WWW - Frontie: Author Contributions Funding Con Ict of Interest Referencestili_secara127No ratings yet
- Biochemistry Free Easy 1Document453 pagesBiochemistry Free Easy 1priyaprasad367792100% (5)
- Easter Revision Day 3 Workbook New 1712407998Document40 pagesEaster Revision Day 3 Workbook New 1712407998robinsonbryNo ratings yet
- Written Report Bio 150Document7 pagesWritten Report Bio 150hyebibieNo ratings yet
- Lippincotts Illustrated Biochem Review 4th Ed Compiled QuestionsDocument36 pagesLippincotts Illustrated Biochem Review 4th Ed Compiled QuestionsKathleen Tubera0% (1)
- MBR 2019 - Biochemistry HandoutsDocument66 pagesMBR 2019 - Biochemistry HandoutsEiaNo ratings yet
- 1 GCE Biology Marking Scheme January 2007 2Document21 pages1 GCE Biology Marking Scheme January 2007 2semirah anthonyNo ratings yet
- Answer Scheme Upp Sem 1 2020 Ssi Kulai Section A (15 Marks) : ConfidentialDocument5 pagesAnswer Scheme Upp Sem 1 2020 Ssi Kulai Section A (15 Marks) : ConfidentialJiaWenTanNo ratings yet
- Anaerobic Respiration PDFDocument7 pagesAnaerobic Respiration PDFmanoj_rkl_07No ratings yet
- Fermentare PDFDocument90 pagesFermentare PDFIstudor AdrianaNo ratings yet
- Metabolisme Protein: Dr. I Dewa Ayu Susilawati, Drg. M. KesDocument31 pagesMetabolisme Protein: Dr. I Dewa Ayu Susilawati, Drg. M. KesMelisa Novitasari100% (2)
- M SC Biotechnology Syllabus&SchemeDocument62 pagesM SC Biotechnology Syllabus&SchemeVaibhav IngoleNo ratings yet