Professional Documents
Culture Documents
1995 - NAR - Solid-Phase Revesible Immobilization For The Isolation of PCR Products
1995 - NAR - Solid-Phase Revesible Immobilization For The Isolation of PCR Products
Uploaded by
Khoa NguyendangOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
1995 - NAR - Solid-Phase Revesible Immobilization For The Isolation of PCR Products
1995 - NAR - Solid-Phase Revesible Immobilization For The Isolation of PCR Products
Uploaded by
Khoa NguyendangCopyright:
Available Formats
4742-4743 Nucleic Acids Research, 1995, Vol. 23, No.
22 1995 Oxford University Press
Solid-phase reversible immobilization for the
isolation of PCR products
Margaret M. DeAngelis, David G. Wang and Trevor L. Hawkins*
Whitehead Institute/MIT, Center for Genome Research, One Kendall Square, Cambridge, MA 02139, USA
Received March 15, 1995; Revised and Accepted October 12, 1995
Large numbers of templates for DNA sequencing can be
produced via PCR directly from plaques, colonies or genomic
DNA. Sequencing directly from PCR products has many
advantages over subcloning; the ability to PCR directly from
plaques or colonies removes the need for template preparation
and is highly amenable to automation. The main problem with
this approach is the subsequent purification of the amplified
products prior to DNA sequencing, especially since the sequence
quality is proportional to the purity of the template. This is
especially important when sequencing PCR products to identify
sequence polymorphisms. Figure 1. This gel shows an example of PCR products before and after
purification using SPRI. Lanes M are 200 ng OX 174 HaeIII digest, lanes I and
The advantages of using magnetic particles in molecular and 2 are one-tenth of a PCR product before and after SPRI purification, lanes 3 and
diagnostic biology have been described previously (14). The use 4 are one tenth of a PCR product spiked with 100 nmol of excess primer (36 nt)
of solid phase techniques has significantly increased over the past prior to purification, lane 3 is before and lane 4 is after SPRI. Lanes 5 and 6 are
few years as more biochemical methods have become adapted for identical to lanes 3 and 4 using a different PCR product. Lanes 7 and 8 are 100
nmol excess primer (36 nt) before and after purification using SPRI.
use with magnetic particles.
We introduce here a general method for producing quality
DNA sequencing template from PCR products. This procedure is
rapid and inexpensive ($0.15 per prep.). The method termed SPRI
(solid-phase reversible immobilization) avoids organic extrac- Standard PCR reaction (50 jil).
tion, filtration and centrifugation steps (5). The SPRI method 1. 6.5 g1 PCR MIX [lOx PCR buffer, 5 ,tl; 10 mM dNTPs, 0.5
employs a carboxyl coated magnetic particle manufactured by ,tl; Taq, 1 U; dH20 to add up to 6.5 g1.
PerSeptive Diagnostics, Cambridge, MA. (cat no #8-4125). We 2. 41 ,ul primer dilution [10 IIM F&R primers, 0.5 ,ul; dH2O,
discovered that these particles could reversibly bind DNA in the 40.5 1l].
presence of polyethylene glycol (PEG) and salt. 3. 2.5 ,ul genomic DNA (50 ng).
This solid phase has no streptavidin, making the use of
biotinylated primers or probes attached to the particles PCR conditions (35 cycles). 96°C, 5 min; 96°C, 30 s; 57°C or
unnecessary. When using biotinylated primers one must exercise 55°C, 2 min; 72°C, 2 min; 72°C, 5 min; 4°C.
caution since excess primer will compete for streptavidin particle
binding (6). This in turn may also contribute to lower yield and Solid-phase reversible immobilization for the purification of PCR
quality of the template. products (96-wellformat).
Here we describe a general PCR isolation procedure which is 1. Wash 10 mg/ml carboxyl coated magnetic particles three
amenable to automation, rapid and yields double-stranded PCR times with WASH BUFFER [0.5 M EDTA (pH 8.0)].
product suitable for DNA sequencing. The method is as follows. 2. For each PCR reaction (50 pul), add 10 pl of washed particles
PCR primers. Forward primers are tailed with -21 M 1 3 and 50 pl of HYB BUFFER (2.5 M NaCl/20% PEG 8000). Mix
sequences. TGTAAAACGACGGCCAGT (18 nt). well and incubate at room temperature for 10 min.
3. Place the microtitre plate on a magnet for 2 min and wash the
PCR reagents. particles twice with 150 p1 of 70% EtOH.
1. lOx PCR buffer [100 mM Tris-HCI (pH 9.3); 500 mM KCI; 4. Air dry for 2 min, and resuspend the particles in 20 pl of
15 mM MgCl2; 0.01% gelatin]. ELUTION BUFFER [ 10 mM Tris-acetate (pH 7.8)] and incubate
2. 10 mM dNTPs. at room temperature for S min.
3. 10 ,tM forward and reverse primers. 5. Magnetically separate the particles and remove the
4. 20 ng/,tl genomic DNA. supematant for testing and sequencing.
*To whom correspondence should be addressed
Nucleic Acids Research, 1995, Vol. 23, No. 22 4743
120 130 140 150 160 170 1e0 190 200 210 220 230
1...........
. I....................I...................I....................I.-.................................I...................I........................................ ................................. .......I.............
ople 31 o'AGCG CCTAACTG
GIG~(ATGGACAG ACMTGGGC7,'A GT3CCCACCC ATAGGrTG0GA TACAAAAGAC AGGCJ&AGGAA GGGrGTAGAAC CATCiAAAGAG GAATAGGC7G1 GTGACCCCAA,
ople 32 ¶1TACGCAGT0Y1 CCTAACTG~GG GGATOGGACAG ACAA¶D30GCA GTG3CCAACCC ATAGGGXGGA TACAAAAGAC AGGCAAGGAA GGGGTAGAAC CATCAAAGAG GAATAGGCTC, GTGACCCCAA
iLple 33 TTAGGCAGTG CCTAACTOGGG GGATGGACAG ACAATGGGCA GTOCCAACCC ATAGGGTGGA TACAAAAGAC AGGCAAGGAA GGGGTAGAAC CATCAAAGAG GAATAGGCTG GTGACCCCAlA
aLple 34 7T1ACGGAGTG CCTAACTGGG GGATOGACAG ACAATGGGCA GTGC-CAACCC ATAGGGLGGA TACAAAAGAC AGGCAAGGAJ GGGGTAGAAC CATCAAAGAG GAATAGGCTG GTGACCCCAA
aLple 35 TTACGCAGTG CCTAACTGGG GGATOGACAG ACAATGGGCA GTGCCAACCC ATAGGGTGGA TACAAAAGAC AGGCAAGGAA GG.GGTAGAAC CA'tAAAGAG GAATAGGCTG GTGACCCCAA
ople 36 T1TACGCAGTG CCTAACTOGG GGATIOGACAG ACAATP3GGCA GTGCCAACCC ATAGGG¶FGGA TACAAAAGAC AGGCAAGGAA GGGGTAGAAC CkOATAAGAG iGATAGGCTG GTGACCCCAA
eple 37 ITTAGGCAGTG CCTAACTGGG GGATGGACAG ACAATCGGGCA GTOCCAACCC ATAGGGTGGA TACAAAAGAC AGGCAAGGAA GGGGTAGAAC CATCAA?&GAG GAATAGGCTO GTGACCCCAA
eple 30 TITAGGCAGTG CCTAACTGGG GGATGGACAG ACAATGGGCA GTGCCAACCC ATAGGGTGGA TACA.AAGAC AGGCAAGGAA CGGGGTAGAAC CA¶IAAAGAG GAATAGGCTG GTGACCCCAA
¶TTAGGCAGTG CCTAACTGGG GGATGGACAG AGGCAAGGAA GGGGTAGAAC CATCAMGAG GAATAGGCTG GTGACCCCAA
U r 5GTGCC-AACCC
kple 39 ACAATOGG(CA ATAGGGTGGA TACAAAAGAC
6Lple 40 rrITCAGCU U 3 n ~ ~ fI h i~ nII U h i U GTGACCCCAA
1471 2750,
114o-i{CCT A CT GC GS3G ATGGAC A GACAATGGG CAGTGC CAAC CC AT A GGCY GOGA T AC AAAA GA CAGGC A A G A A G TA GAAC C ATC AAA
GAOGG A AT A466 CT
1455 2730
1451 27-21
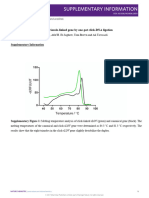
Figure 2. Sequencing traces derived from three related individuals are aligned to show the reliability of calling heterozygous bases. The cycle sequencing reactions
of purified double-stranded SPRI purified PCR products were performed using AmpliTaq ES DNA polymerase (Applied Biosystems Division of Perkin Elmer, CA)
using dye-labeled -21 Ml 3 primers. The reactions were then run on an ABI 373A following the manufacturers protocols.
The SPRI PCR method binds DNA based upon size as shown particles. In addition we would like to thank the members of the
in Figure 1. This figure shows that for a 2 kb PCR product, the DNA Sequencing group for help and support during this project.
final yield is 80-90% whereas the yield from a PCR primer <50 nt This work was supported by NIH and DOE funding.
in length is almost undetectable. We have shown previously that
the lower limit at which yields in excess of 80% are achieved is
200 bp, the maximum limit is in excess of 200 kb (BAC DNA
isolation). REFERENCES
Overall this solid-phase procedure is fast, simple and highly
automatable. Over the past year, this method has been used to 1 Uhlen,M. (1989) Nature 340, 733-744.
isolate >5000 PCR products for DNA sequencing, the majority of 2 Hultman,T., Stahl,S., Hornes,E. and Uhlen,M. (1989) Nucleic Acids Res.
which have been purified on our robotic systems. As shown in 17, 4937-4946.
Figure 2, the sequence data is of the highest quality, allowing the 3 Hawkins,T.L (1992) J. DNA Seq. Mapping 3, 65-69.
identification of single base pair polymorphisms. 4 Hawkins, T.L. (1994) In Craig Venter J. (ed.) Automated DNA sequencing
and analysis.
5 Hawkins,T.L., O'Connor-Morin,T., Roy,A. and Santillan,C. (1994) Nucleic
ACKNOWLEDGEMENTS Acids Res. 22, 4543-4544.
6 Sibson, D.R. (1994) In Craig Venter J. (ed.) Automated DNA sequencing
We would like to thank Dr Chris Bums for supplying the magnetic and analysis.
You might also like
- Hmm202 Practical 1 WorksheetDocument7 pagesHmm202 Practical 1 WorksheetWafaa AdamNo ratings yet
- MatK PCR & Sequencing ProtocolsDocument3 pagesMatK PCR & Sequencing ProtocolsPham Ha Thanh Tung100% (1)
- Amino Acid CodesDocument1 pageAmino Acid Codesmorzan ortizNo ratings yet
- 8.12 RNA Transcription WorksheetDocument2 pages8.12 RNA Transcription WorksheetAmor Reyes EngNo ratings yet
- 8GOMDocument6 pages8GOMaakif inayatNo ratings yet
- Pembimbing: DR - Dr. Mardiastuti H. Wahid, M.SC, SPMK (K)Document27 pagesPembimbing: DR - Dr. Mardiastuti H. Wahid, M.SC, SPMK (K)Hanung PujanggaNo ratings yet
- Table2 PDFDocument1 pageTable2 PDFvaleriaovandoNo ratings yet
- Lab Act 3Document5 pagesLab Act 3Ming VerrierNo ratings yet
- Bacterias Del DesiertoDocument73 pagesBacterias Del DesiertoBrenda ValoNo ratings yet
- pmTFP1 ZyxinDocument5 pagespmTFP1 ZyxinAlleleBiotechNo ratings yet
- pmTFP1 Cx26Document4 pagespmTFP1 Cx26AlleleBiotechNo ratings yet
- pmTFP1 Cx32Document4 pagespmTFP1 Cx32AlleleBiotechNo ratings yet
- pmTFP1 MitochondriaDocument4 pagespmTFP1 MitochondriaAlleleBiotechNo ratings yet
- pmTFP1 ActinDocument4 pagespmTFP1 ActinAlleleBiotechNo ratings yet
- PrimersDocument7 pagesPrimersh gNo ratings yet
- Primer E. ColiDocument4 pagesPrimer E. Coliromario1313No ratings yet
- pmTFP1 FibrillarinDocument4 pagespmTFP1 FibrillarinAlleleBiotechNo ratings yet
- Introduction To The Polymerase Chain Reaction (PCR)Document3 pagesIntroduction To The Polymerase Chain Reaction (PCR)Dr-Dalya ShakirNo ratings yet
- All Ustrious Pmwasabi AnnexinDocument4 pagesAll Ustrious Pmwasabi AnnexinAlleleBiotechNo ratings yet
- Biomol MataDocument64 pagesBiomol MataVita TantiNo ratings yet
- Pmwasabi cx43Document4 pagesPmwasabi cx43AlleleBiotechNo ratings yet
- Supplemental Data Induction of Pluripotent Stem Cells From Mouse Embryonic and Adult Fibroblast Cultures by Defined FactorsDocument41 pagesSupplemental Data Induction of Pluripotent Stem Cells From Mouse Embryonic and Adult Fibroblast Cultures by Defined FactorsewrjdNo ratings yet
- pmTFP1 PeroxisomesDocument4 pagespmTFP1 PeroxisomesAlleleBiotechNo ratings yet
- Cryptoperla Kawasawai Mitochondrial Gene For 16S rRNA, Partial Sequence, Specimen - Voucher: 5386 (Shinshu University, Tojo Laboratory Collection)Document3 pagesCryptoperla Kawasawai Mitochondrial Gene For 16S rRNA, Partial Sequence, Specimen - Voucher: 5386 (Shinshu University, Tojo Laboratory Collection)Diky IgnatiusNo ratings yet
- pmTFP1 ClathrinDocument4 pagespmTFP1 ClathrinAlleleBiotechNo ratings yet
- pmTFP1 VinculinDocument5 pagespmTFP1 VinculinAlleleBiotechNo ratings yet
- 43kda: Rv2276: AccgcgaccgttctgctcgDocument9 pages43kda: Rv2276: AccgcgaccgttctgctcgLeroy WheelerNo ratings yet
- SSR PrimersDocument53 pagesSSR PrimersAlamgir HossainNo ratings yet
- PmTFP1 KeratinDocument4 pagesPmTFP1 KeratinAlleleBiotechNo ratings yet
- Pmwasabi FibrillarinDocument5 pagesPmwasabi FibrillarinAlleleBiotechNo ratings yet
- PaperTrigoHB4Gonzalez Et Al 2019 Fig Compl PDFDocument6 pagesPaperTrigoHB4Gonzalez Et Al 2019 Fig Compl PDFFrancisco AyalaNo ratings yet
- Trower, 1996. in Vitro Mutagenesis Protocols Volume 57Document9 pagesTrower, 1996. in Vitro Mutagenesis Protocols Volume 57afinaNo ratings yet
- PmWasabi KeratinDocument4 pagesPmWasabi KeratinAlleleBiotechNo ratings yet
- Pcr8gwtopo ManDocument44 pagesPcr8gwtopo ManCitlalli CedilloNo ratings yet
- Features TestDocument7 pagesFeatures TestChristophe CullinNo ratings yet
- Bt11 ProtocolDocument6 pagesBt11 ProtocolSonu SomanathNo ratings yet
- All Ustrious Pmwasabi-VinculinDocument5 pagesAll Ustrious Pmwasabi-VinculinAlleleBiotechNo ratings yet
- Assembly of A Biocompatible Triazole-Linked Gene by One-Pot click-DNA Ligation. Suplementary InfomrationDocument15 pagesAssembly of A Biocompatible Triazole-Linked Gene by One-Pot click-DNA Ligation. Suplementary Infomrationdobrovolskis.bioNo ratings yet
- PCR Conditions SummaryDocument5 pagesPCR Conditions Summarygoncalves_alexandreNo ratings yet
- TapanScan Dec 07, 2021Document10 pagesTapanScan Dec 07, 2021Anirban HaldarNo ratings yet
- Tugas Biomol Ardhiyah TsilatsiDocument8 pagesTugas Biomol Ardhiyah TsilatsiArdhiyah DhiyahNo ratings yet
- Pmwasabi TubulinDocument4 pagesPmwasabi TubulinAlleleBiotechNo ratings yet
- pmTFP1 VSV GDocument4 pagespmTFP1 VSV GAlleleBiotechNo ratings yet
- Supplemental Information: Iscience, Volume 26Document9 pagesSupplemental Information: Iscience, Volume 26Ceren CelayirNo ratings yet
- Polymerase Chain Reaction 1Document8 pagesPolymerase Chain Reaction 1Ye Htut AungNo ratings yet
- Lecture 2 DNA Amplification Printable VersionDocument40 pagesLecture 2 DNA Amplification Printable VersionnatnaNo ratings yet
- Mitochondrial Genome Testudinae 2Document7 pagesMitochondrial Genome Testudinae 2JULIO CÉSAR CHÁVEZ GALARZANo ratings yet
- Pmwasabi VimentinDocument4 pagesPmwasabi VimentinAlleleBiotechNo ratings yet
- Perrey 1998Document5 pagesPerrey 1998Shukr Wesman BlbasNo ratings yet
- N32 LCK Sequence AnalysisDocument8 pagesN32 LCK Sequence Analysissrividya84No ratings yet
- Primers SalmonellaDocument1 pagePrimers SalmonellaShamanth RakkithNo ratings yet
- Kaks - Calculator Toolbox 2.0: User'S ManualDocument19 pagesKaks - Calculator Toolbox 2.0: User'S Manual蔡森No ratings yet
- Optimization of The Annealing For DNA Amplification: in VitroDocument4 pagesOptimization of The Annealing For DNA Amplification: in VitroShalini ParthipanNo ratings yet
- Kawazu Et Al 2012 - Supple Transforming Mutations of Rac GTP in Human CancersDocument7 pagesKawazu Et Al 2012 - Supple Transforming Mutations of Rac GTP in Human CancersHernestoNo ratings yet
- Pmwasabi PaxillinDocument4 pagesPmwasabi PaxillinAlleleBiotechNo ratings yet
- Beis47 GGT-LQ 2013 PDFDocument2 pagesBeis47 GGT-LQ 2013 PDFAlejandro MiguelNo ratings yet
- Secuencia de RNAm Maduro y Secuencia DNA (Intrones y Exones) de Proteína p53Document9 pagesSecuencia de RNAm Maduro y Secuencia DNA (Intrones y Exones) de Proteína p53Alejandra Villarreal SánchezNo ratings yet
- Bioinfo Homework 1-1Document24 pagesBioinfo Homework 1-1Rain WinterNo ratings yet
- pmTFP1 VimentinDocument4 pagespmTFP1 VimentinAlleleBiotechNo ratings yet
- Pmwasabi GolgiDocument4 pagesPmwasabi GolgiAlleleBiotechNo ratings yet
- Isolation of Dna From Arthrospira Platensis and Whole Blood Using Magnetic Nanoparticles (Fe O @oa and Fe O @Oa@Sio)Document11 pagesIsolation of Dna From Arthrospira Platensis and Whole Blood Using Magnetic Nanoparticles (Fe O @oa and Fe O @Oa@Sio)Khoa NguyendangNo ratings yet
- Ramli2020 Article PassionFruitPassifloraEdulisPeDocument11 pagesRamli2020 Article PassionFruitPassifloraEdulisPeKhoa NguyendangNo ratings yet
- 2013 - Analytical Biochemistry - One-Stop Genomic DNA Extraction by Salicylic Acid-Coated Magnetic NanoparticlesDocument4 pages2013 - Analytical Biochemistry - One-Stop Genomic DNA Extraction by Salicylic Acid-Coated Magnetic NanoparticlesKhoa NguyendangNo ratings yet
- 2006 - Appl Microbial Biotechnol - Magnetic Partivles For The Seperation and Purification of Nucleic Acids (Mini Review)Document10 pages2006 - Appl Microbial Biotechnol - Magnetic Partivles For The Seperation and Purification of Nucleic Acids (Mini Review)Khoa NguyendangNo ratings yet
- 2006 - Analytical Biochemistry - Application of Magnetic Particles Fe3O4 For Isolation of Genomic DNA From Mammalian CellsDocument3 pages2006 - Analytical Biochemistry - Application of Magnetic Particles Fe3O4 For Isolation of Genomic DNA From Mammalian CellsKhoa NguyendangNo ratings yet
- Replication - Transcription - TranslationDocument75 pagesReplication - Transcription - TranslationJason FryNo ratings yet
- Chemdraw Kimia MedisinalDocument6 pagesChemdraw Kimia MedisinalRisky DaniarNo ratings yet
- Transcription PPTDocument69 pagesTranscription PPTEllieNo ratings yet
- HARPERS - VI Biosynthesis of Nutritionally Nonessential Amino AcidsDocument2 pagesHARPERS - VI Biosynthesis of Nutritionally Nonessential Amino AcidsdandiNo ratings yet
- Periodic Chart of Amino AcidsDocument1 pagePeriodic Chart of Amino AcidsKatherin100% (1)
- Advanced Level Cala - DNA ModelDocument3 pagesAdvanced Level Cala - DNA ModelTafadzwa MachongweNo ratings yet
- Biochem Essential and Non EssentialsDocument2 pagesBiochem Essential and Non EssentialsAlvin Cris RongavillaNo ratings yet
- Dna Structure and FunctionDocument12 pagesDna Structure and FunctionAliah Aziz100% (1)
- Diferensiasi Dan Spesialisasi SelDocument41 pagesDiferensiasi Dan Spesialisasi SelhusnilwardiyahNo ratings yet
- Science: Quarter 3 Module 4 DNA and Protein SynthesisDocument24 pagesScience: Quarter 3 Module 4 DNA and Protein SynthesisJeanne Emerose TalabuconNo ratings yet
- Introduction Too Leo ChemicalsDocument67 pagesIntroduction Too Leo ChemicalsLeni Widiarti LenwidNo ratings yet
- HotStarTaq Plus PCR Master Mix Kit enDocument4 pagesHotStarTaq Plus PCR Master Mix Kit enmutt1190No ratings yet
- Structure of RnaDocument44 pagesStructure of RnaAditi CharakNo ratings yet
- Chapter 11 Test BankDocument11 pagesChapter 11 Test Bankanon_747148947No ratings yet
- Adn ArmableDocument2 pagesAdn ArmableMarcela Ino100% (1)
- GGL DNA Replication PDF VersionDocument7 pagesGGL DNA Replication PDF VersionlloaanaNo ratings yet
- Predominant Forms of Amino AcidsDocument4 pagesPredominant Forms of Amino AcidsAndreaNo ratings yet
- Cold Spring Harb Protoc-2019-Green-pdb - Top095109Document22 pagesCold Spring Harb Protoc-2019-Green-pdb - Top095109FranciscoNo ratings yet
- Assignment Biology (SB 015)Document6 pagesAssignment Biology (SB 015)Wei yangNo ratings yet
- CH8 DNA Structure and Replication 2021Document4 pagesCH8 DNA Structure and Replication 2021EmileMcBrokeNo ratings yet
- Nucleic Acids - DNA and RNADocument53 pagesNucleic Acids - DNA and RNATom Anthony TonguiaNo ratings yet
- Lecture 36 SequencingDocument12 pagesLecture 36 SequencingJenna ScheiblerNo ratings yet
- Biochemistry Review Question: Protein MetabolismDocument13 pagesBiochemistry Review Question: Protein MetabolismrashitaNo ratings yet
- Protein Synthesis: Teacher Answer Key: Code CrackingDocument10 pagesProtein Synthesis: Teacher Answer Key: Code CrackingMariana SuescúnNo ratings yet
- Post Assessment Module 3: Commonly Used Microbes in BiotechnologyDocument4 pagesPost Assessment Module 3: Commonly Used Microbes in BiotechnologyRyan CuisonNo ratings yet
- Chromosomes and DnaDocument5 pagesChromosomes and DnaS. AnsariNo ratings yet
- Exercise and Problems Types of Nucleic Acids (Section 22.1)Document7 pagesExercise and Problems Types of Nucleic Acids (Section 22.1)Azyrah Lyren Seguban UlpindoNo ratings yet
- DNA Replication Worksheet-2 1Document6 pagesDNA Replication Worksheet-2 1La’Niyah WitherspoonNo ratings yet