Professional Documents
Culture Documents
Proton Translocation Pathway: ROS Production (Reviewed in (2) )
Proton Translocation Pathway: ROS Production (Reviewed in (2) )
Uploaded by
050Alya NurOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Proton Translocation Pathway: ROS Production (Reviewed in (2) )
Proton Translocation Pathway: ROS Production (Reviewed in (2) )
Uploaded by
050Alya NurCopyright:
Available Formats
molecule.
An eighth Fe–S cluster, named N1a, located near the FMN, is thought to serve as an electron store
meant
to avoid an excessive ROS production [5].
The quinone-binding site is formed by the 49 kDa-PSST-ND3-ND1 subunits in bacteria [11]. This
module allows electron transfer to the ubiquinone through theN6a and N6b Fe–S clusters located on the TYKY
subunit and through N2 reaction centre cluster.
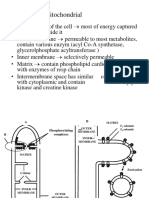
Proton translocation pathway
Four proton channels are located in the membrane domain and participate inprotonpumping: three in antiporter-
like
subunits (ND2, ND4 and ND5) and one at the interface with the hydrophilic domain (ND1-ND6-ND4L) [11].
The
proton-pumping channels through the membrane domain are distant from the redox-active groups mediating
electron
transfer through the hydrophilic domain. The current model for proton pumping proposes that the stabilisation
of a negatively charged ubiquinone at the ubiquinone-binding site would be responsible for a conformational
change
causing energy transmission to the membrane arm and resulting in proton pumping [4,11,14,15].
ROS production (reviewed in [2])
ROS areproduced in two situations: (i) when electrons are backed up in the chain of Fe–S clusters. This happens
when
NADH is present and the downstream chain is inhibited by complex I inhibitors that bind to the quinone-
binding
site or by inhibitors of complex III or complex IV; and (ii) in conditions of reverse electron transfer, when
electrons
are going backwards from complex II via ubiquinone to complex I. In the ‘forward’ mode, the electrons
delivered by
NADH produce ROS at the FMN cofactor; in the ‘reverse mode’, superoxide is generated from a semi-
ubiquinone
at the quinone-binding site. There are also experimental evidences that this mode could also produce superoxide
at
the FMN moiety [16,17] but this is still to be investigated. The reverse electron transfer requires a high
membrane
potential or proton-motive force.
Complex III
Complex III (EC 1.10.2.2), the second proton-pumping complex, catalyses the reaction .
2QH2 + 2 ferricytochrome c + 2H+Matrix → QH2 + 2 ferrocytochrome c + 4H+IMS
Subunit composition
Complex III always functions under a dimeric form, with a size of ≈500 kDa. The monomeric form of complex
III contains ten to eleven subunits: three core subunits of prokaryotic origin, cytochrome b—cytochrome c1—
Rieske iron–sulphur protein, and eight supernumerary subunits. Cytochrome b contains the ubiquinol oxidation
centre (Qo) and ubiquinone reduction centre (Qi), separated by two b-type haems. One haem (bL) is close to the
Qo site and has a low redox potential, and the other haem(bH) is close to the Qi site and has a high potential haem
[17]. Cytochrome c1 contains a c-type haem.
Proton-motive Q cycle and electron transfer
The molecular mechanism that drives H+ pumping of complex III has been first proposed by Mitchell [18] and
supported by numerous studies (reviewed in [2]). In short, the proton-motive Q cycle starts with the oxidation of
ubiquinol at the Qo site, and the release of two H+ at the intermembrane space. One electron is transferred
successively to theFe–S cluster of the Rieske protein, to cytochrome c1, and then to soluble cytochrome c. The
other electron is successively transferred to the haem bL of cytochrome b, to haembH and finally to a ubiquinone
bound to the Qi site, which is reduced to a stabilised semiquinone species. In the second round of the cycle, the
electron entering the Fe–S cluster of the Rieske protein reduces a second cytochrome c and c1, which is
accompanied by two other H+ released at the intermembrane space. The electron entering the low-potential
chain reduces the semiquinone in the Qi site to ubiquinol. This is accompanied by the uptake of two protons
from the negative matrix side of the mitochondrial membrane. Thus there is a net translocation of 2H+/2e−. The
key component of the Q cycle is the correct routing of the two electrons resulting from ubiquinol oxidation to
either the Fe–S cluster of the Rieske protein or to the bL haem of cytochrome b [19,20]. The flexibility of the
globular domain of the Rieske protein seems to be one of the main elements for the bifurcation of the electrons.
Other components implicated in this correct routing could be the binding of ubiquinol into the Q o site that would
trigger conformational changes, maybe due to the widening of Qo to accept the substrate [21].
_c 2018 The Author(s). This is an open access article published by Portland Press Limited on behalf of the Biochemical Society and distributed under the
Creative Commons A
You might also like
- TQMDocument25 pagesTQMAadil Kakar0% (1)
- Oxidative Phosphorylation and Electron Transport Chain II Respiratory Complexes Structure and Organization Chemiosmotic TheoryDocument20 pagesOxidative Phosphorylation and Electron Transport Chain II Respiratory Complexes Structure and Organization Chemiosmotic TheoryIffatnazNo ratings yet
- Electron Transport ChainDocument33 pagesElectron Transport ChainAfaq AhmadNo ratings yet
- BBC2 K27,28RCDocument42 pagesBBC2 K27,28RCMarieta RitongaNo ratings yet
- The Electron Transport System Also Called The Electron Transport ChainDocument5 pagesThe Electron Transport System Also Called The Electron Transport ChainRe UpNo ratings yet
- Electron Transport ChainDocument7 pagesElectron Transport ChainMaria HarisNo ratings yet
- Respiratory Chain & Oxidative PhosphorylationDocument57 pagesRespiratory Chain & Oxidative PhosphorylationHanifa AffianiNo ratings yet
- ASSIGNMENT NO2 ChemDocument5 pagesASSIGNMENT NO2 ChemD AmanatNo ratings yet
- Etc PDFDocument14 pagesEtc PDFjamalNo ratings yet
- Electron Transport Chain ExplainedDocument9 pagesElectron Transport Chain Explainedmaria genioNo ratings yet
- ChemiosmosisDocument7 pagesChemiosmosisSkenzKenzNo ratings yet
- Oxidation Glucose Coenzymes Glycolysis Citric Acid Cycle: Electron TransportDocument4 pagesOxidation Glucose Coenzymes Glycolysis Citric Acid Cycle: Electron TransportHabibur RahamanNo ratings yet
- Gayatree PaniDocument23 pagesGayatree PaniGobindaSahuNo ratings yet
- BiochemDocument10 pagesBiochemHoàng LâmNo ratings yet
- 4 Bioenergetics and Oxidative Metabolism IIDocument3 pages4 Bioenergetics and Oxidative Metabolism IILinus LiuNo ratings yet
- BiologyDocument14 pagesBiologyHarsh SharmaNo ratings yet
- Electron Transport ChainetcDocument19 pagesElectron Transport Chainetcpk kaleenaNo ratings yet
- Electron Transport ChainDocument18 pagesElectron Transport ChainArlyn Pasion BungbongaNo ratings yet
- Oxidative Phosphorylation: Molecular Biochemistry IDocument47 pagesOxidative Phosphorylation: Molecular Biochemistry Iabc007200No ratings yet
- BIO 361 Exam 4 ReviewDocument45 pagesBIO 361 Exam 4 ReviewNigel Zhang100% (1)
- Electron Transport ChainDocument3 pagesElectron Transport ChainEmma MelNo ratings yet
- Biology ProjectDocument16 pagesBiology ProjectSoumya Ranjan PradhanNo ratings yet
- Mitochondria: Mitochondria and Oxidative Phosphorylation Molecular Mechanisms of Electron Transport and Proton PumpingDocument54 pagesMitochondria: Mitochondria and Oxidative Phosphorylation Molecular Mechanisms of Electron Transport and Proton Pumpingnokate konkoorNo ratings yet
- Electron Transport Chain - WikipediaDocument53 pagesElectron Transport Chain - WikipediaLsaurusNo ratings yet
- In Relation To Lsm2101 Lec 2Document11 pagesIn Relation To Lsm2101 Lec 2jojolim18No ratings yet
- Electron Transport SystemDocument58 pagesElectron Transport SystemSantosh KumarNo ratings yet
- Electron Transport Chain: Jump To Navigation Jump To SearchDocument16 pagesElectron Transport Chain: Jump To Navigation Jump To SearchJennie KimNo ratings yet
- Quinone and Non-Quinone Redox Couples in Complex III: # Springer Science + Business Media, LLC 2008Document7 pagesQuinone and Non-Quinone Redox Couples in Complex III: # Springer Science + Business Media, LLC 2008abossyNo ratings yet
- Electron Transport ChainDocument9 pagesElectron Transport Chainalokesh1982100% (1)
- Echevarria Jonille S. BSP 2Document5 pagesEchevarria Jonille S. BSP 2Jonille EchevarriaNo ratings yet
- Chapter 20 ETC and Oxidative PhosphDocument29 pagesChapter 20 ETC and Oxidative PhosphSpencer ThomasNo ratings yet
- Electron Transport and Oxidative Phosphorylation: Refer To: Lehninger Principles of Biochemistry (Chapter 19)Document43 pagesElectron Transport and Oxidative Phosphorylation: Refer To: Lehninger Principles of Biochemistry (Chapter 19)Yousef KhallafNo ratings yet
- Electron Transport ch14Document21 pagesElectron Transport ch14tania.delafuenteNo ratings yet
- Protonmotive Pathways and Mechanisms in The Cytochrome BC ComplexDocument8 pagesProtonmotive Pathways and Mechanisms in The Cytochrome BC ComplexAndrei FloreaNo ratings yet
- Lecture 3 Oxidative Phosphorylation Metabolism IIIDocument32 pagesLecture 3 Oxidative Phosphorylation Metabolism IIIAshish PaswanNo ratings yet
- Aerobic Respiration: Chemiosmosis and Electron Transport ChainDocument17 pagesAerobic Respiration: Chemiosmosis and Electron Transport ChainAihk kenneth BaronaNo ratings yet
- Energy Transducing Membrane: Submitted by - Farheen Khan Roll No - 19Mbs007 Msc. Biosciences 1 Year (2 Semester)Document10 pagesEnergy Transducing Membrane: Submitted by - Farheen Khan Roll No - 19Mbs007 Msc. Biosciences 1 Year (2 Semester)ADITYAROOP PATHAKNo ratings yet
- Biochemical Energy Production Sas PT 3Document5 pagesBiochemical Energy Production Sas PT 3Abegail Ashley PenoniaNo ratings yet
- Regulation of Oxidative PhosphorylationDocument14 pagesRegulation of Oxidative Phosphorylationmaaz629No ratings yet
- Electron Transport Chain - 1Document7 pagesElectron Transport Chain - 1Manash SarmahNo ratings yet
- Biochem Lec32Document5 pagesBiochem Lec32Louis FortunatoNo ratings yet
- LEC 5 Detoxification of Xenobiotics PDFDocument31 pagesLEC 5 Detoxification of Xenobiotics PDFrajeshNo ratings yet
- Biology ReportDocument4 pagesBiology ReportMekailaangela C. EulogioNo ratings yet
- Oxidative Phosphorylation 2022 PDFDocument34 pagesOxidative Phosphorylation 2022 PDFSandy IkbarNo ratings yet
- Electron Transport ChainDocument4 pagesElectron Transport ChainBae SeujiNo ratings yet
- Oxidative Phosphorylation 1Document22 pagesOxidative Phosphorylation 1fatin nadia100% (1)
- Electron TransportDocument8 pagesElectron Transportเทพนิมิตร สมภักดีNo ratings yet
- Chapter 20 Electron Transport and Oxidative PhosphorylationDocument14 pagesChapter 20 Electron Transport and Oxidative PhosphorylationRaabia Ansari100% (1)
- Tutorial 2 NMJ10103 Answer SchemeDocument5 pagesTutorial 2 NMJ10103 Answer SchemeWendy LohNo ratings yet
- CHAPTER 21 PhotosynthesisDocument12 pagesCHAPTER 21 Photosynthesis楊畯凱No ratings yet
- Ch. 9 Biological OxidationDocument71 pagesCh. 9 Biological OxidationKrishna KanthNo ratings yet
- Redox Coupling Electron TransferDocument6 pagesRedox Coupling Electron TransferĐường Ái BìnhNo ratings yet
- Lec-Aeo-11-Biochem 2018Document4 pagesLec-Aeo-11-Biochem 2018mimrahmi7No ratings yet
- Electron Transport Channel & Oxidative PhosphorylationDocument55 pagesElectron Transport Channel & Oxidative PhosphorylationShahabNo ratings yet
- Inhibitors of Mitochondrial Electron Transport-3Document9 pagesInhibitors of Mitochondrial Electron Transport-3ktabbaaNo ratings yet
- Inhibitors of Mitochondrial Electron Transport-2Document9 pagesInhibitors of Mitochondrial Electron Transport-2ktabbaaNo ratings yet
- Electron Transport Chain BSBT025F18 PDFDocument7 pagesElectron Transport Chain BSBT025F18 PDFZaki SyedNo ratings yet
- Electrontransportchain 151027073626 Lva1 App6892Document29 pagesElectrontransportchain 151027073626 Lva1 App6892GobindaSahuNo ratings yet
- Biological OxidationDocument51 pagesBiological OxidationGorav Sharma100% (2)
- Bioelectrochemistry of Biomembranes and Biomimetic MembranesFrom EverandBioelectrochemistry of Biomembranes and Biomimetic MembranesNo ratings yet
- Simulation of Transport in NanodevicesFrom EverandSimulation of Transport in NanodevicesFrançois TriozonNo ratings yet
- Manual de Motor Fuera de Borda Mercury XP 115 ProDocument150 pagesManual de Motor Fuera de Borda Mercury XP 115 ProGestión De MantenimientoNo ratings yet
- Tugas2 M. Ismeth B. INGDocument2 pagesTugas2 M. Ismeth B. INGtugaskls12xpaNo ratings yet
- TEST2Document40 pagesTEST2Kavitha Suresh KumarNo ratings yet
- NCP1Document4 pagesNCP1ejyoung928No ratings yet
- Erik Erickson TheoryDocument4 pagesErik Erickson TheoryMuhammad Ather Siddiqi100% (1)
- Kusudama MeteoroidDocument24 pagesKusudama MeteoroidRaffaela LinetzkyNo ratings yet
- Elementary 1Document4 pagesElementary 1Evelina PalamarciucNo ratings yet
- Laboratory Practical Report: Computer System and Information Technology ApplicationDocument13 pagesLaboratory Practical Report: Computer System and Information Technology ApplicationSantosh RaiNo ratings yet
- ISIELT Lecture 9.2:: Rules of Statutory Interpretation: The Golden RuleDocument12 pagesISIELT Lecture 9.2:: Rules of Statutory Interpretation: The Golden RuleManjare Hassin RaadNo ratings yet
- Technical Interview Study GuideDocument18 pagesTechnical Interview Study GuideRutvij MehtaNo ratings yet
- Sabah Claim According To SalongaDocument17 pagesSabah Claim According To SalongaZoltan Boros100% (1)
- Quantum Hall EffectsDocument131 pagesQuantum Hall EffectsArmando IdarragaNo ratings yet
- 2nd PROF ANIL KUMAR THAKUR MEMORIAL NATIONAL DEBATE COMPETITIONDocument9 pages2nd PROF ANIL KUMAR THAKUR MEMORIAL NATIONAL DEBATE COMPETITIONMovies ArrivalNo ratings yet
- Personal Trading BehaviourDocument5 pagesPersonal Trading Behaviourapi-3739065No ratings yet
- North Jersey Jewish Standard, April 17, 2015Document64 pagesNorth Jersey Jewish Standard, April 17, 2015New Jersey Jewish StandardNo ratings yet
- The Definite Integral and Its ApplicationsDocument13 pagesThe Definite Integral and Its Applicationsapi-312673653100% (1)
- 5G NG RAN Ts 138413v150000p PDFDocument256 pages5G NG RAN Ts 138413v150000p PDFSrinath VasamNo ratings yet
- The Gods Were Astronauts:: Evidence of The True Identities of The Old 'Gods' by Erich Von DänikenDocument44 pagesThe Gods Were Astronauts:: Evidence of The True Identities of The Old 'Gods' by Erich Von DänikenKiran KumarNo ratings yet
- 2232 - Subject - Telecommunication Switching Systems - Year - B.E. (Electronics Telecommunication - Electronics Communication Engineering) Sixth Semester (C.BDocument2 pages2232 - Subject - Telecommunication Switching Systems - Year - B.E. (Electronics Telecommunication - Electronics Communication Engineering) Sixth Semester (C.Bakbarnagani13No ratings yet
- 2017 Catalog PDFDocument60 pages2017 Catalog PDFHasnain KanchwalaNo ratings yet
- Chapter II IntroductionDocument18 pagesChapter II IntroductionMark Jason DuganNo ratings yet
- Assess Skin Turgor and Oral Mucous Membran Es For Signs of Dehydrati On. Note Presence of Nausea, Vomiting and FeverDocument3 pagesAssess Skin Turgor and Oral Mucous Membran Es For Signs of Dehydrati On. Note Presence of Nausea, Vomiting and FeverYamete KudasaiNo ratings yet
- MSAT-BOW-Template - Budget-of-Work-Matatag - Docx 21STDocument15 pagesMSAT-BOW-Template - Budget-of-Work-Matatag - Docx 21STmoanaNo ratings yet
- Autobiography RubricDocument1 pageAutobiography Rubricapi-264460605No ratings yet
- 7.sericulture at RSRSDocument22 pages7.sericulture at RSRSSamuel DavisNo ratings yet
- Cornelius Geoffrey Oslo and MomentDocument51 pagesCornelius Geoffrey Oslo and MomentSarah.MNo ratings yet
- American Government Pre-TestDocument3 pagesAmerican Government Pre-TestMatt HasquinNo ratings yet
- Manager Purchasing Business Development in Lancaster PA Resume Greg ZimmermanDocument2 pagesManager Purchasing Business Development in Lancaster PA Resume Greg ZimmermanGregZimmermanNo ratings yet
- Coolant ConfusionDocument4 pagesCoolant Confusionteodor1974No ratings yet