Professional Documents
Culture Documents
Infographic - CRISPR - Cas9
Infographic - CRISPR - Cas9
Uploaded by
felipe duarteCopyright:
Available Formats
You might also like
- Lab Report CRISPR Sample 1Document9 pagesLab Report CRISPR Sample 1mayagal1707No ratings yet
- MCQ Forensics 1Document29 pagesMCQ Forensics 1Mark Lojero78% (18)
- Rewriting A GenomeDocument2 pagesRewriting A GenomeUjwalNo ratings yet
- CRISPRDocument15 pagesCRISPRserbusabinaNo ratings yet
- Five e Lesson Plan - Dna StructureDocument3 pagesFive e Lesson Plan - Dna Structureapi-336264987No ratings yet
- EDICIÓN GENÉTICA CRISPRcasDocument50 pagesEDICIÓN GENÉTICA CRISPRcasMAKARENA JIMENEZ VILLEGASNo ratings yet
- Crispr Cas 9Document3 pagesCrispr Cas 9E narender nayakNo ratings yet
- Genome Editing and Crispr Cas 9Document12 pagesGenome Editing and Crispr Cas 9HemsaiYadavNo ratings yet
- Protein From Defluviimonas Sр.: Priority Application Title: DNA CUTTING MEANS BASED ON CAS9Document13 pagesProtein From Defluviimonas Sр.: Priority Application Title: DNA CUTTING MEANS BASED ON CAS9MichaelandKaye DanaoNo ratings yet
- Dnipropetrovsk State Medi Cal Academy Unology and Occupational DiseasesDocument66 pagesDnipropetrovsk State Medi Cal Academy Unology and Occupational DiseasesSuba Saravanan 12No ratings yet
- CRISPR Cas9 TechniqueDocument9 pagesCRISPR Cas9 TechniqueAdil ZahoorNo ratings yet
- Genetic Manipulation - Lontoc Aina Gail B.Document2 pagesGenetic Manipulation - Lontoc Aina Gail B.Aina Gail LontocNo ratings yet
- CRISPR/Cas9 & Targeted Genome Editing: New Era in Molecular BiologyDocument7 pagesCRISPR/Cas9 & Targeted Genome Editing: New Era in Molecular BiologykekkaigenkeiNo ratings yet
- Cas 9Document23 pagesCas 9Emey HamdeyNo ratings yet
- Crispr BMB402 2022Document36 pagesCrispr BMB402 2022Mohadev SahaNo ratings yet
- CRISPRDocument15 pagesCRISPRanirbanmanna88320No ratings yet
- Crispr (: / Krɪspər/ DNA Genomes Prokaryotic ArchaeaDocument8 pagesCrispr (: / Krɪspər/ DNA Genomes Prokaryotic ArchaeaIleana StoicaNo ratings yet
- Crispr TechniqueDocument16 pagesCrispr TechniqueAmmar Abbas100% (1)
- CRISPR - Nov27 - 2023 - TaggedDocument55 pagesCRISPR - Nov27 - 2023 - TaggedRitwik DasNo ratings yet
- Prof Silberklang BTECH-480 Term Paper Topic-CRISPR Cas 9 Gene EditingDocument9 pagesProf Silberklang BTECH-480 Term Paper Topic-CRISPR Cas 9 Gene EditingkonarkNo ratings yet
- Crispr 101: Your Guide To Understanding CRISPRDocument29 pagesCrispr 101: Your Guide To Understanding CRISPRydydNo ratings yet
- Crispr Cas9 and Targeted Genome EditingDocument5 pagesCrispr Cas9 and Targeted Genome EditingDaniH46No ratings yet
- Crispr: Presented By: Anurag Chauhan (21513) M.Sc. Microbial-Biotechnology - IV Department of BiotechnologyDocument14 pagesCrispr: Presented By: Anurag Chauhan (21513) M.Sc. Microbial-Biotechnology - IV Department of BiotechnologyAnurag ChauhanNo ratings yet
- Animal Biotechnology: Theory AssignmentDocument14 pagesAnimal Biotechnology: Theory AssignmentISHIKA TYAGINo ratings yet
- CRISPR EbookDocument14 pagesCRISPR EbookBayan MallahNo ratings yet
- North South University: CRISPR-Cas9Document5 pagesNorth South University: CRISPR-Cas9ishita hossainNo ratings yet
- Crispr Week 12-13 by AyeshaDocument12 pagesCrispr Week 12-13 by AyeshaAyesha KhalidNo ratings yet
- Wolter & Putcha (2018) - Plant Transcription FactorsDocument18 pagesWolter & Putcha (2018) - Plant Transcription FactorsAna Luiza Atella de FreitasNo ratings yet
- Ma 2014Document20 pagesMa 2014hiwmacrigeeeNo ratings yet
- Crispr: PHR 423 Nusrat HossainDocument16 pagesCrispr: PHR 423 Nusrat HossainMd. Minhazul IslamNo ratings yet
- Addgene CRISPR GuideDocument18 pagesAddgene CRISPR GuideThomasNo ratings yet
- Gabr Mahmoud ThesisDocument35 pagesGabr Mahmoud Thesismahmoud gabrNo ratings yet
- Applications of CRISPR-Cas For Synthetic Biology and Genetic RecordingDocument7 pagesApplications of CRISPR-Cas For Synthetic Biology and Genetic RecordingShampa SenNo ratings yet
- ReportDocument6 pagesReportramna ziaNo ratings yet
- The FEBS Journal - 2014 - Ma - Genome Modification by CRISPR Cas9Document8 pagesThe FEBS Journal - 2014 - Ma - Genome Modification by CRISPR Cas9Alberto Luis Lizcano GonzálezNo ratings yet
- NEB Featurearticle Crispr Cas9 Summer2014 LowresDocument6 pagesNEB Featurearticle Crispr Cas9 Summer2014 LowresRakhee RachelNo ratings yet
- Crispr 101: Your Guide To Understanding CRISPRDocument19 pagesCrispr 101: Your Guide To Understanding CRISPRYassier AnwarNo ratings yet
- CRISPR/Cas9 For Genome Editing: Progress, Implications and ChallengesDocument7 pagesCRISPR/Cas9 For Genome Editing: Progress, Implications and ChallengesAngelica RestrepoNo ratings yet
- Research Problem Assignment Week 6Document3 pagesResearch Problem Assignment Week 6Pearlyn LauNo ratings yet
- Cas9 As A Versatile Tool For Engineering Biology: PerspectiveDocument7 pagesCas9 As A Versatile Tool For Engineering Biology: PerspectiveLetícia AlibertiNo ratings yet
- CRISPR/Cas9 Immune System As A Tool For Genome EngineeringDocument8 pagesCRISPR/Cas9 Immune System As A Tool For Genome EngineeringmariaNo ratings yet
- Supplementary Materials For: RNA-Guided Human Genome Engineering Via Cas9Document36 pagesSupplementary Materials For: RNA-Guided Human Genome Engineering Via Cas9Ronilo Jose Danila FloresNo ratings yet
- CRISPR1Document7 pagesCRISPR1cesarNo ratings yet
- 816 (2012) Martin Jinek: Science Et AlDocument7 pages816 (2012) Martin Jinek: Science Et AlDarko PopovskiNo ratings yet
- 2021-Review-Single-Base Resolution Increasing The Specificity of The CRISPR-Cas SystemDocument12 pages2021-Review-Single-Base Resolution Increasing The Specificity of The CRISPR-Cas SystemCristian Felipe Sandoval QuiñonezNo ratings yet
- ZFN, Talens, Crispr PDFDocument9 pagesZFN, Talens, Crispr PDFjezelle lividNo ratings yet
- Past, Present, and Future of CRISPR Genome Editing TechnologiesDocument25 pagesPast, Present, and Future of CRISPR Genome Editing TechnologiesZiyaNo ratings yet
- Jennifer Doudna CRISPR SystemDocument14 pagesJennifer Doudna CRISPR SystemLightspeedNo ratings yet
- Crispr Cas9 PDFDocument16 pagesCrispr Cas9 PDFIves LucasNo ratings yet
- 12 MolecularBiologytechniquesCRISPR-CasSystemDocument8 pages12 MolecularBiologytechniquesCRISPR-CasSystemSusovan SadhukhanNo ratings yet
- Koonin, Makarova, 2013Document8 pagesKoonin, Makarova, 2013gerardrianaceaeNo ratings yet
- CRISPR Genome Editing and Its Medical ApplicationsDocument8 pagesCRISPR Genome Editing and Its Medical ApplicationsAn ex-prodigy YT junkie named FreshNo ratings yet
- CRISPR 101 Ebook PDFDocument29 pagesCRISPR 101 Ebook PDFMarco Antonio Diaz GonzalezNo ratings yet
- Crispr 101: Your Guide To Understanding CRISPRDocument29 pagesCrispr 101: Your Guide To Understanding CRISPRPradipta BNo ratings yet
- Synthego Ebook Crispr 101 PDFDocument29 pagesSynthego Ebook Crispr 101 PDFRie R100% (1)
- Review: The Biology of CRISPR-Cas: Backward and ForwardDocument21 pagesReview: The Biology of CRISPR-Cas: Backward and ForwardNiharika ChaudharyNo ratings yet
- Advances in CRISPR-Cas9 Genome Engineering: Lessons Learned From RNA InterferenceDocument13 pagesAdvances in CRISPR-Cas9 Genome Engineering: Lessons Learned From RNA InterferenceAmina Tucak-SmajićNo ratings yet
- (CRISPR) Literature Review RaissaDocument11 pages(CRISPR) Literature Review RaissaRaissa E. FedoraNo ratings yet
- CRISPR+101+eBook v2021Document20 pagesCRISPR+101+eBook v2021Ruan GonçalvesNo ratings yet
- Crispr Cas HajarDocument21 pagesCrispr Cas HajarHajira Fatima100% (1)
- CrisprDocument6 pagesCrispranuradhaclNo ratings yet
- SMJ 64 88Document2 pagesSMJ 64 88felipe duarteNo ratings yet
- Infografía Determining Breast Cancer Biomarker Status and Associated Morphological Features Using Deep LearningDocument3 pagesInfografía Determining Breast Cancer Biomarker Status and Associated Morphological Features Using Deep Learningfelipe duarteNo ratings yet
- Therapeutic Targeting of Angiogenesis Molecular Pathways in Angiogenesis-Dependent DiseasesDocument11 pagesTherapeutic Targeting of Angiogenesis Molecular Pathways in Angiogenesis-Dependent Diseasesfelipe duarteNo ratings yet
- Infographic Polycystic Ovary Syndrome Definition, Aetiology, Diagnosis and TreatmentDocument4 pagesInfographic Polycystic Ovary Syndrome Definition, Aetiology, Diagnosis and Treatmentfelipe duarteNo ratings yet
- Summary - The Endoplasmic Reticulum Stress Response in Disease Pathogenesis and PathophysiologyDocument4 pagesSummary - The Endoplasmic Reticulum Stress Response in Disease Pathogenesis and Pathophysiologyfelipe duarteNo ratings yet
- Botany Syllabus For PG Assistant (Source Internet)Document4 pagesBotany Syllabus For PG Assistant (Source Internet)Selvaraju ParthibhanNo ratings yet
- F. Brock Fuller - Decomposition of The Linking Number of A Closed Ribbon: A Problem From Molecular BiologyDocument5 pagesF. Brock Fuller - Decomposition of The Linking Number of A Closed Ribbon: A Problem From Molecular BiologyDopameNo ratings yet
- Science 10 3rd Quarter Exam: Share This DocumentDocument1 pageScience 10 3rd Quarter Exam: Share This DocumentJAYM DELOS SANTOSNo ratings yet
- PDF Reference2-Lesson10-DnaandchromosomesDocument3 pagesPDF Reference2-Lesson10-Dnaandchromosomesjg teNo ratings yet
- Dna Polymerase: Classification of Dna PolymerasesDocument8 pagesDna Polymerase: Classification of Dna PolymerasesBOMMIDI JAHNAVI (RA2132001010057)No ratings yet
- METALLODRUGSDocument21 pagesMETALLODRUGSAditya Verma100% (1)
- Di truyền cơ sở-Ch11 (14) - Genetic Analysis and Biotechnology-IUHDocument80 pagesDi truyền cơ sở-Ch11 (14) - Genetic Analysis and Biotechnology-IUHNam NguyenHoangNo ratings yet
- Human Genome Project Fridovich 2023Document6 pagesHuman Genome Project Fridovich 2023Santiago Castro HernándezNo ratings yet
- SR Elite, Aiims S60 & Neet MPL Aits Grand Test - 15 Paper Key (23-04-2023)Document12 pagesSR Elite, Aiims S60 & Neet MPL Aits Grand Test - 15 Paper Key (23-04-2023)vulurakashsharma2005No ratings yet
- Joseph Michael Cabaya General Biology Lesson 5: Evolution and Origin of Biodiversity: Patterns of Descent With ModificationDocument5 pagesJoseph Michael Cabaya General Biology Lesson 5: Evolution and Origin of Biodiversity: Patterns of Descent With ModificationJulimar CabayaNo ratings yet
- AmaR One PCRDocument2 pagesAmaR One PCRWang EvanNo ratings yet
- A 2.3 HL VirusesDocument25 pagesA 2.3 HL Virusestaiga.aisaka14404No ratings yet
- Aryan HatwarDocument15 pagesAryan HatwarAryan HatwarNo ratings yet
- VCE Biology Study Design 2013 - 2016Document50 pagesVCE Biology Study Design 2013 - 2016SarahGeorgieNo ratings yet
- Hatonn, Gyeorgos - The Last Great PlagueDocument152 pagesHatonn, Gyeorgos - The Last Great PlagueKanaubaNo ratings yet
- Transcription and RegulationDocument40 pagesTranscription and RegulationRUDRANSH PUJARINo ratings yet
- Application of Recombinant DNA TechnologyDocument24 pagesApplication of Recombinant DNA TechnologyMuhammad Husnain WarraichNo ratings yet
- Dna Model 2013 - 2 PDFDocument2 pagesDna Model 2013 - 2 PDFDun TaringaNo ratings yet
- Powerpoint Pembelajaran Bahasa Inggris SMA Kelas XII 2018Document89 pagesPowerpoint Pembelajaran Bahasa Inggris SMA Kelas XII 2018Johana Wulandari75% (8)
- 2022 Specimen Paper 4 Mark SchemeDocument20 pages2022 Specimen Paper 4 Mark Schemeadel hanyNo ratings yet
- Construction of cDNA LibraryDocument7 pagesConstruction of cDNA LibraryTapasya Changkakati100% (1)
- Organisation and Control of Prokaryotic and Eukaryotic GenomeDocument8 pagesOrganisation and Control of Prokaryotic and Eukaryotic GenomeNicholas OwNo ratings yet
- Arber and LinnDocument7 pagesArber and Linnshubham20No ratings yet
- Odyssey High School Biology VocabularyDocument24 pagesOdyssey High School Biology VocabularyHanQian WangNo ratings yet
- Hannah Ness - Mutations Webquest - 3071914Document5 pagesHannah Ness - Mutations Webquest - 3071914Hannah NessNo ratings yet
- ExcerptDocument10 pagesExcerptAlifah RizkaNo ratings yet
- La Biología de La Cromatina Complejos de Remodelación: FurtherDocument24 pagesLa Biología de La Cromatina Complejos de Remodelación: FurtherVíc AltamarNo ratings yet
- Rings (Also Known As The Holy Trinity), Though There Are OthersDocument92 pagesRings (Also Known As The Holy Trinity), Though There Are OthersOnur Mutlu100% (2)
Infographic - CRISPR - Cas9
Infographic - CRISPR - Cas9
Uploaded by
felipe duarteOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Infographic - CRISPR - Cas9
Infographic - CRISPR - Cas9
Uploaded by
felipe duarteCopyright:
Available Formats
CRISPR/CAS9
FROM BACTERIAL ADAPTIVE IMMUNE SYSTEM TO GENE
EDITING (1)
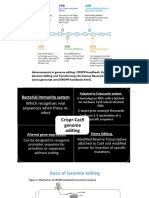
It was discovered that bacteria had repetitive DNA sequences in their genomes called
CRISPRs (Clusters of regularly interspaced short palindromic repeats).
In bacterial chromosomes there were found repetitive sequences of DNA (see black
diamonds in figure 1) that flank unique sequences (see different color squares) that come
from viral DNA. CRISPR segments are repetitive sequences in bacterial DNA separated by
short segments called spacer DNA (similar to virus DNA sequence). Near CRISPRs, Cas
genes were close.
Figure 1. Adapted from (2)
How does it works in bacteria to protect
them from viruses?
When a virus infects a bacteria (and if the bacteria
survives to the infection), the bacteria can acquire
segments of viral DNA in its own genome forming CRISPR
arrays. Fragments of invading foreign DNA integrate within
the host CRISPR locus, these integrated fragments are
called spacer DNA and are responsible for the "adaptive
immunity". So, when another infection begins, the bacteria
use the arrays to transcribe them into a single strand of
RNA molecule. This strand of RNA broke into smaller units
that are composed of a sequence derived from viral DNA
together with another sequence derived from bacterial
DNA adjacent to the viral DNA in the CRISPR array.
These fragments of RNA bind tracrRNA (Tracr binds to the
bacterial DNA repeat derived RNA sequence). These two
molecules of RNA form a structure to bind the enzyme
Cas9. These ARN-protein complex surveys the cell looking
for viral DNA sequences that match the sequence of the
complex ARN (derived from the DNA viral sequence in
CRISPRs). When a sequence matches, Cas 9 unwinds the
infectious viral DNA to allow RNA/DNA hybridization and a
double strand broke is induced in the infectious viral DNA
and its degradation prevents virions formations.
The events are represented in figure 2.
Note: The CRISPR RNA (crRNA) is composed of two parts:
1. The spacer derived RNA. So RNA guide sequences are
derived from the spacer DNA sequences.
2. And RNA derived from the repeat sequences of
bacterial DNA in CRISPR loci in bacterial genome.
Cas proteins along with other RNA components (like
TracrRNA) recognise crRNA due to the presence of the
RNA sequence derived from the bacterial DNA repeated
sequences in CRISPR loci.
The crRNA recognizes complementary sequences in
invading DNA and base pair with these molecules, which
initiates cleavage of the foreign DNA molecule.
Types of systems
CRISPR systems are classified according to the numbers
and types of Cas proteins found in the systems.
Class I system include multiple genes and multiple Cas
proteins. And are more difficult to harness these protein
for gene editing.
Class II system include a single genes encoding one large
protein that combines with CRISPR ARNs. Are easy to
harness. This system is the one that uses TracrRNA that
binds CRISPR ARN to form a single guide form to Cas9 to
cut. The natural CRISPR ARN (the one derived from from
bacterial DNA adjacent to the viral DNA in the CRISPR Figure 2. Adapted from (2)
array) and TracrRNA are combined linked together
covalently making a single transcript.
The systems only differ in the mechanism that produces
CRISPR RNA and Cas proteins.
Cas9 protein
Is an enzyme that recognise double stranded
DNA at positions in the DNA sequence that
match 20 nucleotide sequence in the single
stranded ARN guide. And makes double
strand DNA cuts.
It unwinds DNA without ATP or GTP use. This
is done by conformational changes when
binding to nucleic acids.
HNH domain is one of the chemical clivers of
DNA. And it is thought that the non target
strand of DNA must be present in the
complex in order to achieve cutting. Figure 2. Adapted from (2)
The protein itself is a sensor of the degree of
the complementary match in guideRNA-
targedDNA sequences. When nucleotide
Why and how it is used for gene
complementarity is full the enzyme completely editing
activates and cuts the double strand DNA
target. It is very simple to design single strained RNA
guide molecule, all needed to know is which one is
Mutations in Cas 9 can make the protein a the DNA target sequence and by Watson-Crick
better sensor in order to be more accurate in base pairing rules, a complementary RNA single
cutting and gene editing. string is designed.
In order to cut precisely the desired DNA Every time it is used the same protein (Cas9), so it
molecule recognition must occur between the is no needed to design new proteins each time it is
RNA guide and DNA target, but only if the going to be used.
target DNA sequence is next to a protospacer
adjacent motif (PAM) that are close to the The CRISPR(Cas9 system is only used to guide
recognition sequence in the DNA target Cas9 to acquire desirable double strand cuts in
molecule. specific sequences of DNA, further that point of
the double break everything that happens
depends on cellular DNA repair activities.
Gene editing can help to cure diseased caused
Types of systems only by mutations in the DNA sequence, like sickle
cell anemia
CRISPR systems are classified according to
the numbers and types of Cas proteins Subsequent DNA repair
found in the systems.
In homologous recombination repair (HDR)
Class I system include multiple genes and mechanism, a donor DNA that is provided by the
multiple Cas proteins. And are more difficult experimenter a(that has the modifications wanted
to harness these protein for gene editing. for the homologous sequence in cell target DNA),
serve as a template for repair for the double
Class II system include a single genes strand break, and sequence repair then could
encoding one large protein that combines include the modifications wanted by the
with CRISPR ARNs. Are easy to harness. This experimenter. (3)
system is the one that uses TracrRNA that It is important that the donor sequence for
binds CRISPR ARN to form a single guide homologous repair has light changes on it, in
form to Cas9 to cut. order to prevent re cut by Cas9 complex, changes
in the recognition sequences for example
Note: TracrRNA is transcribed from (mutations in PAMs could work).
upstream of CRISPR loci from genomic loci. the ends of the break start searching for
And Cas9 belongs to system type II. homology, either in a sister chromatid (like in
recombination in meiosis), or in an homologous
chromosome, or look for the donor DNA
homologous sequence.
so is needed to have homology in the donor to al
least one side of the break (minimum for things to
Anti-CRISPRS work fine).
This anti-CRISPRS can inhibit the CRISPR Cas Non homologous join repair (NHEJ) mechanism
pathway, by stopping CRISPR proteins, prevent could be used by cells, and it is a panic response
binding to RNA, etc. Produced by phages, have (there is not an homologous sequence to use as a
genes that encode this inhibitory proteins (very template), because the cell prefers to jam in the
small, aprox 100 aa). ends of the broke DNA rather than lose the
complete segment. and this end direct joining
C1 is one example that blocks the DNA cutting. could make mistakes, like localized insertions and
They can be used to limit activity of CRISPR Cas, to deletions, and produce targeted mutations (small
reduce activities that may be occurring at off- insertions and occasionally larger insertions. And
target sites. deletions with microhomologies that can be
recognized because the sequence that is at the
junction could be assigned either to the left side or
the right side of the junction). Deletions that are in
coding sequence could be an in frame deletions or
out or frame deletions depending if the number of
nucleotides deleted are multiple of 3 (in frame
deletions) or are not multiple of 3 (out of frame
Other forms of gene editing deletions). (3)
Zinc‐finger nucleases (ZFNs) and
transcription activator‐like effector
nucleases (TALENs) are other examples of
genome editing machines. That make
double strain DNA break. But CRISPR/Cas9
is way more simple, cheaper and effective.
Aplications
Genome wide CRISPR
With CRISPR/Cas9 technology, it can be produced knockout (by
libraries
NHEJ), knockin (by HDR) and knock off animal models. As well it
can be generated site specific mutagenesis (by NHEJ) and There are three of them:
corrections (by HDR). 1. CRISPR knockout. Is a loss of function
The system of gene editing is cheap, easy to manage, has a lot method, identifies new biological
of success and it is applicable to every form of living cell with mechanism.
DNA genome. 2. CRISPRa activation. A gene activation
approach ised in screening the gain of
In cancer can be used to enhance ex vivo lymphocytes that are function.
more efficient killing tumor cells once introduced again in the 3. CRISPRi inhibition. A gene inhibition
subject. Or can be used to generate mutation in cancer cells in that is used in screening the gost for
order to kill them. Can help to find knew immunotherapy two main loss of functions.
targets for cancer and other diseases.
It can be used to cure illnesses produced only by DNA
mutations. And can be a knew way to treat and cure
neurodegenerative diseases.
Can be used to alter the HIV genome and inactivate expression
of HIV genes, and make immune cells to the HIV infections.
It can be used as an antimicrobial agent, to kill antibiotic
resistant bacterial strains. It can cleave antibiotic resistant
genes. And in agriculture can be useful too.
The SHERLOCK technique is developed with CRISPR Cas, it
consist of identification of specific RNA or DNA sequences by
using fluorescent molecules attached to Cas9.
And helps in the development of synthetic biological circuits.
REFERENCES
1. Khanzadi MN, Khan AA. CRISPR/Cas9: Nature’s gift to prokaryotes and an
auspicious tool in genome editing. J Basic Microbiol. 2020;60(2):91–102.
2. Jennifer Doudna: CRISPR Basics - YouTube [Internet]. [cited 2021 Apr 4].
Available from: https://www.youtube.com/watch?v=47pkFey3CZ0
3. Dana Carroll: Background on Genome Editing - YouTube [Internet]. [cited 2021
Apr 3]. Available from: https://www.youtube.com/watch?v=upzSmpnxNig
MADE BY : JUAN FELIPE DUARTE ZAMBRANO
MAIL: JDUARTEZ @ UNAL EDU CO. .
04/04/2021
You might also like
- Lab Report CRISPR Sample 1Document9 pagesLab Report CRISPR Sample 1mayagal1707No ratings yet
- MCQ Forensics 1Document29 pagesMCQ Forensics 1Mark Lojero78% (18)
- Rewriting A GenomeDocument2 pagesRewriting A GenomeUjwalNo ratings yet
- CRISPRDocument15 pagesCRISPRserbusabinaNo ratings yet
- Five e Lesson Plan - Dna StructureDocument3 pagesFive e Lesson Plan - Dna Structureapi-336264987No ratings yet
- EDICIÓN GENÉTICA CRISPRcasDocument50 pagesEDICIÓN GENÉTICA CRISPRcasMAKARENA JIMENEZ VILLEGASNo ratings yet
- Crispr Cas 9Document3 pagesCrispr Cas 9E narender nayakNo ratings yet
- Genome Editing and Crispr Cas 9Document12 pagesGenome Editing and Crispr Cas 9HemsaiYadavNo ratings yet
- Protein From Defluviimonas Sр.: Priority Application Title: DNA CUTTING MEANS BASED ON CAS9Document13 pagesProtein From Defluviimonas Sр.: Priority Application Title: DNA CUTTING MEANS BASED ON CAS9MichaelandKaye DanaoNo ratings yet
- Dnipropetrovsk State Medi Cal Academy Unology and Occupational DiseasesDocument66 pagesDnipropetrovsk State Medi Cal Academy Unology and Occupational DiseasesSuba Saravanan 12No ratings yet
- CRISPR Cas9 TechniqueDocument9 pagesCRISPR Cas9 TechniqueAdil ZahoorNo ratings yet
- Genetic Manipulation - Lontoc Aina Gail B.Document2 pagesGenetic Manipulation - Lontoc Aina Gail B.Aina Gail LontocNo ratings yet
- CRISPR/Cas9 & Targeted Genome Editing: New Era in Molecular BiologyDocument7 pagesCRISPR/Cas9 & Targeted Genome Editing: New Era in Molecular BiologykekkaigenkeiNo ratings yet
- Cas 9Document23 pagesCas 9Emey HamdeyNo ratings yet
- Crispr BMB402 2022Document36 pagesCrispr BMB402 2022Mohadev SahaNo ratings yet
- CRISPRDocument15 pagesCRISPRanirbanmanna88320No ratings yet
- Crispr (: / Krɪspər/ DNA Genomes Prokaryotic ArchaeaDocument8 pagesCrispr (: / Krɪspər/ DNA Genomes Prokaryotic ArchaeaIleana StoicaNo ratings yet
- Crispr TechniqueDocument16 pagesCrispr TechniqueAmmar Abbas100% (1)
- CRISPR - Nov27 - 2023 - TaggedDocument55 pagesCRISPR - Nov27 - 2023 - TaggedRitwik DasNo ratings yet
- Prof Silberklang BTECH-480 Term Paper Topic-CRISPR Cas 9 Gene EditingDocument9 pagesProf Silberklang BTECH-480 Term Paper Topic-CRISPR Cas 9 Gene EditingkonarkNo ratings yet
- Crispr 101: Your Guide To Understanding CRISPRDocument29 pagesCrispr 101: Your Guide To Understanding CRISPRydydNo ratings yet
- Crispr Cas9 and Targeted Genome EditingDocument5 pagesCrispr Cas9 and Targeted Genome EditingDaniH46No ratings yet
- Crispr: Presented By: Anurag Chauhan (21513) M.Sc. Microbial-Biotechnology - IV Department of BiotechnologyDocument14 pagesCrispr: Presented By: Anurag Chauhan (21513) M.Sc. Microbial-Biotechnology - IV Department of BiotechnologyAnurag ChauhanNo ratings yet
- Animal Biotechnology: Theory AssignmentDocument14 pagesAnimal Biotechnology: Theory AssignmentISHIKA TYAGINo ratings yet
- CRISPR EbookDocument14 pagesCRISPR EbookBayan MallahNo ratings yet
- North South University: CRISPR-Cas9Document5 pagesNorth South University: CRISPR-Cas9ishita hossainNo ratings yet
- Crispr Week 12-13 by AyeshaDocument12 pagesCrispr Week 12-13 by AyeshaAyesha KhalidNo ratings yet
- Wolter & Putcha (2018) - Plant Transcription FactorsDocument18 pagesWolter & Putcha (2018) - Plant Transcription FactorsAna Luiza Atella de FreitasNo ratings yet
- Ma 2014Document20 pagesMa 2014hiwmacrigeeeNo ratings yet
- Crispr: PHR 423 Nusrat HossainDocument16 pagesCrispr: PHR 423 Nusrat HossainMd. Minhazul IslamNo ratings yet
- Addgene CRISPR GuideDocument18 pagesAddgene CRISPR GuideThomasNo ratings yet
- Gabr Mahmoud ThesisDocument35 pagesGabr Mahmoud Thesismahmoud gabrNo ratings yet
- Applications of CRISPR-Cas For Synthetic Biology and Genetic RecordingDocument7 pagesApplications of CRISPR-Cas For Synthetic Biology and Genetic RecordingShampa SenNo ratings yet
- ReportDocument6 pagesReportramna ziaNo ratings yet
- The FEBS Journal - 2014 - Ma - Genome Modification by CRISPR Cas9Document8 pagesThe FEBS Journal - 2014 - Ma - Genome Modification by CRISPR Cas9Alberto Luis Lizcano GonzálezNo ratings yet
- NEB Featurearticle Crispr Cas9 Summer2014 LowresDocument6 pagesNEB Featurearticle Crispr Cas9 Summer2014 LowresRakhee RachelNo ratings yet
- Crispr 101: Your Guide To Understanding CRISPRDocument19 pagesCrispr 101: Your Guide To Understanding CRISPRYassier AnwarNo ratings yet
- CRISPR/Cas9 For Genome Editing: Progress, Implications and ChallengesDocument7 pagesCRISPR/Cas9 For Genome Editing: Progress, Implications and ChallengesAngelica RestrepoNo ratings yet
- Research Problem Assignment Week 6Document3 pagesResearch Problem Assignment Week 6Pearlyn LauNo ratings yet
- Cas9 As A Versatile Tool For Engineering Biology: PerspectiveDocument7 pagesCas9 As A Versatile Tool For Engineering Biology: PerspectiveLetícia AlibertiNo ratings yet
- CRISPR/Cas9 Immune System As A Tool For Genome EngineeringDocument8 pagesCRISPR/Cas9 Immune System As A Tool For Genome EngineeringmariaNo ratings yet
- Supplementary Materials For: RNA-Guided Human Genome Engineering Via Cas9Document36 pagesSupplementary Materials For: RNA-Guided Human Genome Engineering Via Cas9Ronilo Jose Danila FloresNo ratings yet
- CRISPR1Document7 pagesCRISPR1cesarNo ratings yet
- 816 (2012) Martin Jinek: Science Et AlDocument7 pages816 (2012) Martin Jinek: Science Et AlDarko PopovskiNo ratings yet
- 2021-Review-Single-Base Resolution Increasing The Specificity of The CRISPR-Cas SystemDocument12 pages2021-Review-Single-Base Resolution Increasing The Specificity of The CRISPR-Cas SystemCristian Felipe Sandoval QuiñonezNo ratings yet
- ZFN, Talens, Crispr PDFDocument9 pagesZFN, Talens, Crispr PDFjezelle lividNo ratings yet
- Past, Present, and Future of CRISPR Genome Editing TechnologiesDocument25 pagesPast, Present, and Future of CRISPR Genome Editing TechnologiesZiyaNo ratings yet
- Jennifer Doudna CRISPR SystemDocument14 pagesJennifer Doudna CRISPR SystemLightspeedNo ratings yet
- Crispr Cas9 PDFDocument16 pagesCrispr Cas9 PDFIves LucasNo ratings yet
- 12 MolecularBiologytechniquesCRISPR-CasSystemDocument8 pages12 MolecularBiologytechniquesCRISPR-CasSystemSusovan SadhukhanNo ratings yet
- Koonin, Makarova, 2013Document8 pagesKoonin, Makarova, 2013gerardrianaceaeNo ratings yet
- CRISPR Genome Editing and Its Medical ApplicationsDocument8 pagesCRISPR Genome Editing and Its Medical ApplicationsAn ex-prodigy YT junkie named FreshNo ratings yet
- CRISPR 101 Ebook PDFDocument29 pagesCRISPR 101 Ebook PDFMarco Antonio Diaz GonzalezNo ratings yet
- Crispr 101: Your Guide To Understanding CRISPRDocument29 pagesCrispr 101: Your Guide To Understanding CRISPRPradipta BNo ratings yet
- Synthego Ebook Crispr 101 PDFDocument29 pagesSynthego Ebook Crispr 101 PDFRie R100% (1)
- Review: The Biology of CRISPR-Cas: Backward and ForwardDocument21 pagesReview: The Biology of CRISPR-Cas: Backward and ForwardNiharika ChaudharyNo ratings yet
- Advances in CRISPR-Cas9 Genome Engineering: Lessons Learned From RNA InterferenceDocument13 pagesAdvances in CRISPR-Cas9 Genome Engineering: Lessons Learned From RNA InterferenceAmina Tucak-SmajićNo ratings yet
- (CRISPR) Literature Review RaissaDocument11 pages(CRISPR) Literature Review RaissaRaissa E. FedoraNo ratings yet
- CRISPR+101+eBook v2021Document20 pagesCRISPR+101+eBook v2021Ruan GonçalvesNo ratings yet
- Crispr Cas HajarDocument21 pagesCrispr Cas HajarHajira Fatima100% (1)
- CrisprDocument6 pagesCrispranuradhaclNo ratings yet
- SMJ 64 88Document2 pagesSMJ 64 88felipe duarteNo ratings yet
- Infografía Determining Breast Cancer Biomarker Status and Associated Morphological Features Using Deep LearningDocument3 pagesInfografía Determining Breast Cancer Biomarker Status and Associated Morphological Features Using Deep Learningfelipe duarteNo ratings yet
- Therapeutic Targeting of Angiogenesis Molecular Pathways in Angiogenesis-Dependent DiseasesDocument11 pagesTherapeutic Targeting of Angiogenesis Molecular Pathways in Angiogenesis-Dependent Diseasesfelipe duarteNo ratings yet
- Infographic Polycystic Ovary Syndrome Definition, Aetiology, Diagnosis and TreatmentDocument4 pagesInfographic Polycystic Ovary Syndrome Definition, Aetiology, Diagnosis and Treatmentfelipe duarteNo ratings yet
- Summary - The Endoplasmic Reticulum Stress Response in Disease Pathogenesis and PathophysiologyDocument4 pagesSummary - The Endoplasmic Reticulum Stress Response in Disease Pathogenesis and Pathophysiologyfelipe duarteNo ratings yet
- Botany Syllabus For PG Assistant (Source Internet)Document4 pagesBotany Syllabus For PG Assistant (Source Internet)Selvaraju ParthibhanNo ratings yet
- F. Brock Fuller - Decomposition of The Linking Number of A Closed Ribbon: A Problem From Molecular BiologyDocument5 pagesF. Brock Fuller - Decomposition of The Linking Number of A Closed Ribbon: A Problem From Molecular BiologyDopameNo ratings yet
- Science 10 3rd Quarter Exam: Share This DocumentDocument1 pageScience 10 3rd Quarter Exam: Share This DocumentJAYM DELOS SANTOSNo ratings yet
- PDF Reference2-Lesson10-DnaandchromosomesDocument3 pagesPDF Reference2-Lesson10-Dnaandchromosomesjg teNo ratings yet
- Dna Polymerase: Classification of Dna PolymerasesDocument8 pagesDna Polymerase: Classification of Dna PolymerasesBOMMIDI JAHNAVI (RA2132001010057)No ratings yet
- METALLODRUGSDocument21 pagesMETALLODRUGSAditya Verma100% (1)
- Di truyền cơ sở-Ch11 (14) - Genetic Analysis and Biotechnology-IUHDocument80 pagesDi truyền cơ sở-Ch11 (14) - Genetic Analysis and Biotechnology-IUHNam NguyenHoangNo ratings yet
- Human Genome Project Fridovich 2023Document6 pagesHuman Genome Project Fridovich 2023Santiago Castro HernándezNo ratings yet
- SR Elite, Aiims S60 & Neet MPL Aits Grand Test - 15 Paper Key (23-04-2023)Document12 pagesSR Elite, Aiims S60 & Neet MPL Aits Grand Test - 15 Paper Key (23-04-2023)vulurakashsharma2005No ratings yet
- Joseph Michael Cabaya General Biology Lesson 5: Evolution and Origin of Biodiversity: Patterns of Descent With ModificationDocument5 pagesJoseph Michael Cabaya General Biology Lesson 5: Evolution and Origin of Biodiversity: Patterns of Descent With ModificationJulimar CabayaNo ratings yet
- AmaR One PCRDocument2 pagesAmaR One PCRWang EvanNo ratings yet
- A 2.3 HL VirusesDocument25 pagesA 2.3 HL Virusestaiga.aisaka14404No ratings yet
- Aryan HatwarDocument15 pagesAryan HatwarAryan HatwarNo ratings yet
- VCE Biology Study Design 2013 - 2016Document50 pagesVCE Biology Study Design 2013 - 2016SarahGeorgieNo ratings yet
- Hatonn, Gyeorgos - The Last Great PlagueDocument152 pagesHatonn, Gyeorgos - The Last Great PlagueKanaubaNo ratings yet
- Transcription and RegulationDocument40 pagesTranscription and RegulationRUDRANSH PUJARINo ratings yet
- Application of Recombinant DNA TechnologyDocument24 pagesApplication of Recombinant DNA TechnologyMuhammad Husnain WarraichNo ratings yet
- Dna Model 2013 - 2 PDFDocument2 pagesDna Model 2013 - 2 PDFDun TaringaNo ratings yet
- Powerpoint Pembelajaran Bahasa Inggris SMA Kelas XII 2018Document89 pagesPowerpoint Pembelajaran Bahasa Inggris SMA Kelas XII 2018Johana Wulandari75% (8)
- 2022 Specimen Paper 4 Mark SchemeDocument20 pages2022 Specimen Paper 4 Mark Schemeadel hanyNo ratings yet
- Construction of cDNA LibraryDocument7 pagesConstruction of cDNA LibraryTapasya Changkakati100% (1)
- Organisation and Control of Prokaryotic and Eukaryotic GenomeDocument8 pagesOrganisation and Control of Prokaryotic and Eukaryotic GenomeNicholas OwNo ratings yet
- Arber and LinnDocument7 pagesArber and Linnshubham20No ratings yet
- Odyssey High School Biology VocabularyDocument24 pagesOdyssey High School Biology VocabularyHanQian WangNo ratings yet
- Hannah Ness - Mutations Webquest - 3071914Document5 pagesHannah Ness - Mutations Webquest - 3071914Hannah NessNo ratings yet
- ExcerptDocument10 pagesExcerptAlifah RizkaNo ratings yet
- La Biología de La Cromatina Complejos de Remodelación: FurtherDocument24 pagesLa Biología de La Cromatina Complejos de Remodelación: FurtherVíc AltamarNo ratings yet
- Rings (Also Known As The Holy Trinity), Though There Are OthersDocument92 pagesRings (Also Known As The Holy Trinity), Though There Are OthersOnur Mutlu100% (2)