Professional Documents
Culture Documents
Seminar: Mitchell R Lunn, Ching H Wang
Seminar: Mitchell R Lunn, Ching H Wang
Uploaded by
Catalina AlzogarayCopyright:
Available Formats
You might also like
- Neurology Multiple Choice Questions With Explanations: Volume IFrom EverandNeurology Multiple Choice Questions With Explanations: Volume IRating: 4 out of 5 stars4/5 (7)
- Alternative SplicingDocument356 pagesAlternative SplicingChalitha somarathne100% (3)
- Fast Facts: Diagnosing Amyotrophic Lateral Sclerosis: Clinical wisdom to facilitate faster diagnosisFrom EverandFast Facts: Diagnosing Amyotrophic Lateral Sclerosis: Clinical wisdom to facilitate faster diagnosisNo ratings yet
- ATROFIA MUSCULAR ESPINAL Lancet 2008Document14 pagesATROFIA MUSCULAR ESPINAL Lancet 2008Sebastián Silva SotoNo ratings yet
- Case Report: Coexistence of Ankylosing Spondylitis and Neurofibromatosis Type 1Document4 pagesCase Report: Coexistence of Ankylosing Spondylitis and Neurofibromatosis Type 1Lupita Mora LobacoNo ratings yet
- Benefits of Robotic Devices in Medical Rehabilitation of A Case With Neurofibromatosis Type1Document5 pagesBenefits of Robotic Devices in Medical Rehabilitation of A Case With Neurofibromatosis Type1Cioara IleanaNo ratings yet
- Congenital Muscular Dystrophies and Congenital.8 PDFDocument26 pagesCongenital Muscular Dystrophies and Congenital.8 PDFlithium2203No ratings yet
- Intensive Care Unit-Acquired WeaknessDocument13 pagesIntensive Care Unit-Acquired WeaknessAnonymous 3VkEwIsXRNo ratings yet
- Myoclonus With DementiaDocument8 pagesMyoclonus With DementiaSantosh DashNo ratings yet
- Amyotrophic Lateral SclerosisDocument11 pagesAmyotrophic Lateral SclerosisAnomalie12345No ratings yet
- Romero 2013Document16 pagesRomero 2013Rizki Nandasari SulbahriNo ratings yet
- A Review On Spinal Muscular Atrophy Clinical Classification, Etiology, Diagnosis and TreatmentDocument4 pagesA Review On Spinal Muscular Atrophy Clinical Classification, Etiology, Diagnosis and TreatmentEditor IJTSRDNo ratings yet
- Isolated Propriospinal Myoclonus As A Presentation of Cervical Myelopathy.Document2 pagesIsolated Propriospinal Myoclonus As A Presentation of Cervical Myelopathy.arturomarticarvajalNo ratings yet
- 2 - Debilidad AgudaDocument15 pages2 - Debilidad AgudaMarian ZeaNo ratings yet
- (2022) - Frozen Shoulder ReviewDocument16 pages(2022) - Frozen Shoulder ReviewCristobal LopezNo ratings yet
- Neuromuscular DisordersDocument12 pagesNeuromuscular DisordersZed HarrisNo ratings yet
- Transverse Myelitis: Clinical PracticeDocument9 pagesTransverse Myelitis: Clinical PracticearnabNo ratings yet
- Spinal Muscular Atrophy: Stephen J. Kolb,, John T. KisselDocument16 pagesSpinal Muscular Atrophy: Stephen J. Kolb,, John T. KisselZouhour MiladiNo ratings yet
- A Review of Lumbar Radiculopathy, Diagnosis, and TreatmentDocument7 pagesA Review of Lumbar Radiculopathy, Diagnosis, and TreatmentMatthew PhillipsNo ratings yet
- Evaluating Lumbar Spine 582-589Document8 pagesEvaluating Lumbar Spine 582-589Mansour ZamaniNo ratings yet
- Amyotrophic Lateral SclerosisDocument9 pagesAmyotrophic Lateral SclerosisCaroline ItnerNo ratings yet
- Jurnal SpondylosisDocument7 pagesJurnal SpondylosisM Hafiz MoulviNo ratings yet
- Neuromuscular Diseases PDFDocument205 pagesNeuromuscular Diseases PDFsalmazzNo ratings yet
- Osteochondroses: Franklin Danger, MD Christopher Wasyliw, MD Laura Varich, MDDocument7 pagesOsteochondroses: Franklin Danger, MD Christopher Wasyliw, MD Laura Varich, MDadrianNo ratings yet
- Hercules Baby - A Rare Case Presentation of Congenital MyopathyDocument4 pagesHercules Baby - A Rare Case Presentation of Congenital MyopathyInternational Journal of Innovative Science and Research TechnologyNo ratings yet
- En A04v86n4Document10 pagesEn A04v86n4Florentina NastaseNo ratings yet
- RRL - Scoliosis - Etiologic StudiesDocument6 pagesRRL - Scoliosis - Etiologic StudiesShao Anunciacion0% (1)
- Ogino and Wilson 2004Document16 pagesOgino and Wilson 2004akshayajainaNo ratings yet
- Case Study Myasthenia GravisDocument9 pagesCase Study Myasthenia GravisYow Mabalot100% (1)
- CV 20231223 32653A702 HonbunDocument7 pagesCV 20231223 32653A702 HonbunGrc hashtagNo ratings yet
- Preferred Examination: Spondylitis Ankylosing SpondylitisDocument6 pagesPreferred Examination: Spondylitis Ankylosing SpondylitischrisantyNo ratings yet
- A Pattern Recognition Approach To Myopathy.15Document24 pagesA Pattern Recognition Approach To Myopathy.15Maryam NabNo ratings yet
- Algoritmo Diferencial de La Ataxia Sensitiva en Mielopatia Por Cobre Secundaria A CeliaquiaDocument7 pagesAlgoritmo Diferencial de La Ataxia Sensitiva en Mielopatia Por Cobre Secundaria A CeliaquiaFarid Santiago Abedrabbo LombeydaNo ratings yet
- Algoritmo Diferencial de Debilidad Proximal en Paciente Con Miopatia Adquirida Por Nemalina.Document7 pagesAlgoritmo Diferencial de Debilidad Proximal en Paciente Con Miopatia Adquirida Por Nemalina.Farid Santiago Abedrabbo LombeydaNo ratings yet
- Clinical and Pathologic Aspects of Congenital Myopathies: Ikuya Nonaka MDDocument8 pagesClinical and Pathologic Aspects of Congenital Myopathies: Ikuya Nonaka MDEvlin KoharNo ratings yet
- Cervical SpondylosisDocument10 pagesCervical Spondylosistaufiq duppa duppaaNo ratings yet
- Marinesco-Sjo Gren Syndrome - 2013Document5 pagesMarinesco-Sjo Gren Syndrome - 2013Irfan RazaNo ratings yet
- Tob 12.2Document28 pagesTob 12.2LT DRAGONXNo ratings yet
- Ankylosing Spondylarthritis PDFDocument3 pagesAnkylosing Spondylarthritis PDFMa OlayaNo ratings yet
- Congenital Myopathies: Clinical Phenotypes and New Diagnostic ToolsDocument16 pagesCongenital Myopathies: Clinical Phenotypes and New Diagnostic ToolsRizki Nandasari SulbahriNo ratings yet
- Miopati - Referensi MiopatiDocument25 pagesMiopati - Referensi MiopatiKelvin Theandro GotamaNo ratings yet
- Fas CaseDocument2 pagesFas Caseanon_378668477No ratings yet
- Arthrogryposis Causes, Consequences and Clinical Course in Amyoplasia and Distal ArthrogryposisDocument64 pagesArthrogryposis Causes, Consequences and Clinical Course in Amyoplasia and Distal ArthrogryposisJanina DrăgoiNo ratings yet
- Cervical Spondylosis and Neck PainDocument5 pagesCervical Spondylosis and Neck PainIsaac AlemanNo ratings yet
- Care PlanDocument20 pagesCare PlanamantelNo ratings yet
- Atrfoia Espinal MioclonicDocument18 pagesAtrfoia Espinal MioclonicmmblozsnoNo ratings yet
- Protrusio Acetabuli Diagnosis and TreatmentDocument10 pagesProtrusio Acetabuli Diagnosis and TreatmentFerney Leon BalceroNo ratings yet
- Spinal Muscular AtrophyDocument11 pagesSpinal Muscular AtrophyRahma NovitasariNo ratings yet
- Case Report of Intrafamilial Variability in Autosomal Recessive Centronuclear Myopathy Associated To A Novel BIN1 Stop MutationDocument6 pagesCase Report of Intrafamilial Variability in Autosomal Recessive Centronuclear Myopathy Associated To A Novel BIN1 Stop Mutationyael1991No ratings yet
- Maedica Art 18Document6 pagesMaedica Art 18Ersya MuslihNo ratings yet
- 03 Sarah OConnorDocument3 pages03 Sarah OConnorMateen ShukriNo ratings yet
- A Review of Lumbar Radiculopathy, Diagnosis, and TreatmentDocument7 pagesA Review of Lumbar Radiculopathy, Diagnosis, and TreatmentZuraidaNo ratings yet
- Distrofias Musculares 2019 MercuriDocument14 pagesDistrofias Musculares 2019 Mercurimaria palaciosNo ratings yet
- Idiopathic ScoliosisDocument12 pagesIdiopathic ScoliosisAlessandro BergantinNo ratings yet
- Spinal Muscular AthropyDocument13 pagesSpinal Muscular AthropyooccacNo ratings yet
- DownloadDocument15 pagesDownloadM Aqil GibranNo ratings yet
- (Nigel G. Laing) The Sarcomere and Skeletal Muscle PDFDocument246 pages(Nigel G. Laing) The Sarcomere and Skeletal Muscle PDFVladimir Pablo Morales QuevedoNo ratings yet
- Autoimmune and Paraneoplastic MyeloDocument12 pagesAutoimmune and Paraneoplastic MyeloSimply SamNo ratings yet
- Debilidad Neuromuscular Adquirida y Movilizacion Temprana en UTIDocument14 pagesDebilidad Neuromuscular Adquirida y Movilizacion Temprana en UTILucho FelicevichNo ratings yet
- HTTPS:WWW Ncbi NLM Nih gov:pmc:articles:PMC9144471:pdf:jpm-12-00808Document12 pagesHTTPS:WWW Ncbi NLM Nih gov:pmc:articles:PMC9144471:pdf:jpm-12-00808Ihsanul Ma'arifNo ratings yet
- Journal of Neurology and NeurocienceDocument49 pagesJournal of Neurology and NeurocienceInternational Medical PublisherNo ratings yet
- Genic-Balance Theory of Sex DeterminationDocument19 pagesGenic-Balance Theory of Sex Determinationstevensb05575% (4)
- Bioinformatic PracticeDocument4 pagesBioinformatic Practiceyungiang157No ratings yet
- 1 s2.0 S1044579X20302790 Main PDFDocument10 pages1 s2.0 S1044579X20302790 Main PDFgarrobosNo ratings yet
- Gene ConceptDocument80 pagesGene ConceptPrince HamdaniNo ratings yet
- How The Genes Are Regulated & Expressed?: AssignmentDocument17 pagesHow The Genes Are Regulated & Expressed?: AssignmentSuraiya Siraj NehaNo ratings yet
- Molecular Biology of The Cell, Sixth Edition Chapter 7: Control of Gene ExpressionDocument36 pagesMolecular Biology of The Cell, Sixth Edition Chapter 7: Control of Gene ExpressionIsmael Torres-Pizarro100% (2)
- 002 - 2018 - Splicing Mutations in Human Genetic Disorders - Examples Detection and ConfirmationDocument16 pages002 - 2018 - Splicing Mutations in Human Genetic Disorders - Examples Detection and ConfirmationAndrea Bermúdez QuinteroNo ratings yet
- In Silico Analysis of BRCA1 andDocument23 pagesIn Silico Analysis of BRCA1 and02213No ratings yet
- Unit-3 BioinformaticsDocument15 pagesUnit-3 Bioinformaticsp vmuraliNo ratings yet
- RNA Processing in Eukaryotes: January 15, 2021 Gaurab KarkiDocument5 pagesRNA Processing in Eukaryotes: January 15, 2021 Gaurab KarkiShahriar ShamimNo ratings yet
- Review Article: Hereditary Breast Cancer: The Era of New Susceptibility GenesDocument12 pagesReview Article: Hereditary Breast Cancer: The Era of New Susceptibility GenesSara MagoNo ratings yet
- Transcription Sample Old Essay PDFDocument7 pagesTranscription Sample Old Essay PDFOccamsRazorNo ratings yet
- Mutaciones BGM PDFDocument6 pagesMutaciones BGM PDFwaldo tapiaNo ratings yet
- Primer Design Using IDT Quest: Identifying NCBI Transcript & Associated PrimersDocument3 pagesPrimer Design Using IDT Quest: Identifying NCBI Transcript & Associated PrimersAqsa MuzammilNo ratings yet
- Han 2010Document14 pagesHan 2010julia rossoNo ratings yet
- Lesson Plan Day 1Document3 pagesLesson Plan Day 1Stephanie Ngooi Ming MingNo ratings yet
- Scientific Report 2007: 3D Glasses InsideDocument38 pagesScientific Report 2007: 3D Glasses Insidea4agarwal100% (5)
- Unit 2 Cells, Development, Biodiversity and ConservationDocument47 pagesUnit 2 Cells, Development, Biodiversity and ConservationEllane leeNo ratings yet
- 357 - Cell-Biology Physiology) DNA Transcription001Document12 pages357 - Cell-Biology Physiology) DNA Transcription001SodysserNo ratings yet
- News and Views: Advancing RNA-Seq AnalysisDocument3 pagesNews and Views: Advancing RNA-Seq AnalysisWilson Rodrigo Cruz FlorNo ratings yet
- Vanilloid (Capsaicin) Receptors in Health and Disease: Arpad Szallasi, MD, PHDDocument12 pagesVanilloid (Capsaicin) Receptors in Health and Disease: Arpad Szallasi, MD, PHDprift2No ratings yet
- Data RetrievalDocument17 pagesData RetrievalAyesha Khan50% (2)
- AV UWorld EOs (Rough Draft) - Data - Subject LandscapeDocument139 pagesAV UWorld EOs (Rough Draft) - Data - Subject Landscapesaeedfadaly1No ratings yet
- Gene Expression in EukaryotesDocument26 pagesGene Expression in EukaryotesArap DomNo ratings yet
- Myth of Junk DNA Notes (50p)Document50 pagesMyth of Junk DNA Notes (50p)siriusaz9398100% (1)
- Plant Grafting: PrimerDocument6 pagesPlant Grafting: Primerikhlas65No ratings yet
- Intelligence N GenesDocument208 pagesIntelligence N Genestoussman80No ratings yet
- Comparative Proteomics Kit I: Protein Profiler Module: Biotechnology ExplorerDocument50 pagesComparative Proteomics Kit I: Protein Profiler Module: Biotechnology ExplorerHidayat FadelanNo ratings yet
- RNA SplicingDocument8 pagesRNA SplicingDivya NarayanNo ratings yet
Seminar: Mitchell R Lunn, Ching H Wang
Seminar: Mitchell R Lunn, Ching H Wang
Uploaded by
Catalina AlzogarayOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Seminar: Mitchell R Lunn, Ching H Wang
Seminar: Mitchell R Lunn, Ching H Wang
Uploaded by
Catalina AlzogarayCopyright:
Available Formats
Seminar
Spinal muscular atrophy
Mitchell R Lunn, Ching H Wang
Lancet 2008; 371: 2120–33 Spinal muscular atrophy is an autosomal recessive neurodegenerative disease characterised by degeneration of spinal
Department of Medicine, cord motor neurons, atrophy of skeletal muscles, and generalised weakness. It is caused by homozygous disruption
Stanford University School of of the survival motor neuron 1 (SMN1) gene by deletion, conversion, or mutation. Although no medical treatment is
Medicine, Stanford, CA, USA
(M R Lunn BS); and Department
available, investigations have elucidated possible mechanisms underlying the molecular pathogenesis of the disease.
of Neurology and Neurological Treatment strategies have been developed to use the unique genomic structure of the SMN1 gene region. Several
Sciences, Pediatric Neurology candidate treatment agents have been identified and are in various stages of development. These and other advances
Division, Stanford University in medical technology have changed the standard of care for patients with spinal muscular atrophy. In this Seminar,
Medical Center, Stanford, CA,
USA (C H Wang MD)
we provide a comprehensive review that integrates clinical manifestations, molecular pathogenesis, diagnostic
strategy, therapeutic development, and evidence from clinical trials.
Correspondence to:
Dr Ching H Wang, Department of
Neurology and Neurological Introduction paralysis, and no control of head movement. They are
Sciences, Pediatric Neurology Spinal muscular atrophy, the leading genetic cause of unable to sit without support. The spared diaphragm,
Division, Stanford University
Medical Center, 300 Pasteur
infant deaths, is an autosomal recessive disease that results combined with weakened intercostal muscles, results in
Drive, Room A343, Stanford, from degeneration of motor neurons of the spinal cord. paradoxical breathing (inward bony thorax movement
CA 94305-5235, USA The incidence of spinal muscular atrophy is about one in with outward abdominal movement during inspiration)
wangch@stanford.edu 10 000 livebirths with a carrier frequency of one in 50.1,2 and a bell-shaped upper torso. Bulbar denervation results
There is no effective medical treatment for spinal muscular in tongue fasciculation and weakness with poor suck and
atrophy. However, since the discovery of the disease-causing swallow. It also decreases airway protection and increases
gene—survival motor neuron 1 (SMN1)—substantial the risk of aspiration pneumonia, an important cause of
progress in understanding the molecular pathogenesis of morbidity and mortality.
this disease has been made. With this knowledge, Type II disease is of intermediate severity and
innovations have generated excitement about the imminent characterised by onset between 7 and 18 months of age.
development of treatment options. These and other Patients are able to maintain a sitting position unaided. A
advancements in medical technology have changed the few are able to stand with leg braces, but none can walk
standard of care for patients with this debilitating disease. independently. Kyphoscoliosis usually develops, requiring
surgical or orthotic intervention. Fine tremors with digit
Clinical manifestations extension or hand grips are also common. Weak
Spinal muscular atrophy is characterised by degeneration swallowing might deter weight gain. Like patients with
of motor neurons of the spinal cord, which results in type I disease, clearing of tracheal secretions and coughing
hypotonia and muscle weakness. Previously, diagnosis might become difficult because of poor bulbar function
was confirmed by electromyography and muscle biopsy. and weak intercostal muscles. Respiratory insufficiency is
Electromyography characteristic of the disorder shows a frequent cause of death during adolescence.
features of denervation with spontaneous activity of Patients with type III spinal muscular atrophy
positive sharp waves, fibrillation, and occasional (Kugelberg-Welander disease) show profound symptom
fasciculations.3,4 Motor unit action potential shows high heterogeneity. They typically reach all major motor
amplitudes and long durations coupled with decreased milestones, such as independent walking. Some might
recruitment. Histopathology of skeletal muscle usually need wheelchair assistance in childhood, whereas others
shows atrophic fibres with islands of group hypertrophy,3 might continue to walk and live productive adult lives
and the spinal cord shows severe loss of motor neuron in with minor muscular weakness. These patients often
the anterior horn region (figure 1). develop scoliosis. Symptoms of joint overuse, generally
Spinal muscular atrophy was previously divided into caused by weakness, are frequently seen. Patients with
three clinical types on the basis of age of onset and motor type IV disease typically have onset of weakness in the
function achieved: (1) severe type I; (2) intermediate type
II; and (3) mild type III.5 Adult-onset type IV has been
added to include very mild disease (table 1).6,7 Most Search strategy and selection criteria
researchers define spinal muscular atrophy type by the We searched for all publications containing “spinal muscular
highest level of motor function—ie, sitter, walker. atrophy”, “SMA”, “survival motor neuron”, and “survival of
Type I disease (Werdnig-Hoffmann disease) is the most motor neuron”. We included all relevant papers after 2000,
severe and common type, which accounts for about but seminal reports, regardless of their publication date, were
50% of patients diagnosed with spinal muscular atrophy.8 also included. We reviewed papers published in English,
It is distinguished by the onset of disease before 6 months German, Italian, Japanese, Russian, and Spanish, but included
of age and death within the first 2 years of life. These only those published in English.
patients have profound hypotonia, symmetrical flaccid
2120 www.thelancet.com Vol 371 June 21, 2008
Seminar
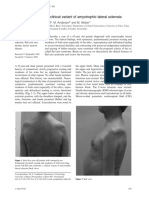
second or third decade of life. Motor impairment is mild
without respiratory or nutritional problems, and patients A B
are able to walk in adult years. Other forms of spinal
muscular atrophy include X-linked disease, spinal
muscular atrophy with respiratory distress (SMARD),
spinal and bulbar muscular atrophy (Kennedy’s disease),
and distal spinal muscular atrophy, which are beyond the
scope of this Seminar.
Normal SMA
Molecular genetics and pathogenesis
In 1990, investigators used linkage analysis to identify
the locus of the SMA gene on chromosome 5q13.9,10 The
initial 10 cM interval was later narrowed to a 1–2 cM
region with recombinant mapping.11–13 Cloning of the
disease-gene region with yeast artificial chromosomes
identified multicopy repeats of microsatellites (eg, CMS1,
C212, C272) and other genomic sequences (figure 2).
This unstable region was subjected to intrachromosomal
rearrangements, including gene duplications, gene C D
conversions, and de-novo deletions.14–16 This complex
genomic organisation greatly hindered the initial pro-
gress of cloning the disease gene.11,15 In 1994, researchers *
identified deletions of multicopy microsatellites in
patients with spinal muscular atrophy;15 some of which
were in linkage disequilibrium with the disease locus.17
Despite the genomic complexity in this region, Melki and Normal SMA
colleagues15 uncovered a small 11 kb fragment that
uniquely hybridised to a telomeric repeat that was Figure 1: Histopathology of spinal muscular atrophy
Motor commands generated in the cerebral cortex are transmitted through spinal cord α-motor neurons (red cell
missing in patients, thereby further narrowing the in spinal cord anterior horn and green arrow in (A). The spinal cord anterior horn region shows an absence of
candidate region. motor neurons in a patient (B) compared with those in the healthy control (A). Skeletal muscle of a patient (D)
A human fetal brain cDNA library was probed by shows hypertrophic fibres (hollow arrowhead) surrounded by group atrophy (green arrowhead) compared with
genomic DNA from the candidate region, and the SMN healthy fibres with uniform morphology in normal infantile muscle (C). Despite the atrophy of muscle fibres in
spinal muscular atrophy, muscle spindles (black asterisk) are not affected and become more conspicuous (D). All
gene was identified as the disease-causing gene in 1995.18 slides are stained with haematoxylin and eosin.
The SMN gene is present in multiple copies in the human
genome: one SMN1 (SMNT, telomeric) and several SMN2 OMIM Age at onset Highest function Natural age
(SMNC, centromeric). The two genes differ by only five number achieved of death
nucleotides, only one of which is within the 1·7 kb coding Type I (severe, Werdnig-Hoffmann disease) 253300 0–6 months Never sit <2 years
region, but none affect the predicted aminoacid Type II (intermediate) 253550 7–18 months Sit, never stand >2 years
sequence.18,19 Both genes contain nine exons and eight Type III (mild, Kugelberg-Welander disease) 253400 >18 months Stand and walk Adult
introns that span about 20 kb genomic region. SMN1 Type IV (adult) 271150 Second or third Walk during Adult
encodes a 38 kDa protein with 294 aminoacids. The SMN decade adulthood
protein is expressed in all somatic tissues and is highly
OMIM=Online Mendelian Inheritance in Man.
conserved from yeast to man.20–22 The SMN gene duplication
found in primates took place after the split of primates Table 1: Classification criteria for spinal muscular atrophy
from rodents, since mice have only one copy, which is
denoted Smn. SMN2, however, is unique to man because no exon seven (figure 3).25,26 The resultant SMNΔ7 protein
chimpanzees do not have this gene signature despite is non-functional and thought to be rapidly degraded.25,27,28
having multiple SMN gene copies in their genome.23 About 10% of SMN2 pre-mRNA is properly spliced and
More than 98% of patients with spinal muscular subsequently translated into full-length SMN protein.18
atrophy have a homozygous disruption of SMN1 by However, the efficiency of SMN2 splicing might be
deletion, rearrangement, or mutation.18,24 All these dependent on severity of disease, and production of a
patients, however, retain at least one copy of SMN2. This full-length transcript of SMN2 could range from 10%
gene undergoes alternative splicing and produces a to 50%.29 This low level of SMN protein allows embryonic
truncated mRNA isoform in which exon seven (major development, but is not sufficient to sustain the survival
product) or exon five, or both, are absent. A C-to-T of motor neurons of the spinal cord.
nucleotide transition at position six of exon seven (Ex7+6) In 2002, Cartegni and Kranier30 reported that the C-to-T
is responsible for the alternatively spliced isoform with nucleotide transition at Ex7+6 in SMN2 disrupts the
www.thelancet.com Vol 371 June 21, 2008 2121
Seminar
Centromeric copy Telomeric copy
C272 C272
CATT–1 CMS1 CATT–1 C171 AG1–CA C212 C212 AG1–CA C171 CATT–1 CMS1
Cen Tel
GTF2H2 NAIP SMN2 SERF1 SERF1 SMN1 NAIP GTF2H2
SMA genetic map on chromosome 5q13
Figure 2: Genetic map of the spinal muscular atrophy locus
SMN gene copies are contained within two large inverted genomic fragments (black horizontal lines) within this region on chromosome 5q13. SMN1 (blue) is located
within the telomeric copy whereas at least one SMN2 (blue) is contained within the centromeric copy. Other genes in the vicinity of SMN copies are shown in black.
Blue and red arrows denote the direction of transcription of SMN and other genes, respectively. Multicopy microsatellite markers (red) are indicated within these
genomic repeats.
SR
SR
SF2 Tra2
U2 U2
ASF 100%
1
PA
SMN1 6 snRNP AF ESE 7 8 (A)n 6 7 8 (A)n
RN
hn
CAGACAA
~10%
SR
Tra2
~90%
RN 1
hn P A1
1
hn P A
PA
PA
SMN2 6 ESE 7 8 (A)n 6 8 (A)n
RN
RN
RN
hn
hn
U2 U2
snRNP AF
UAGACAA
Figure 3: Pre-mRNA splicing of SMN1 and SMN2
In SMN1, an exonic splicing enhancer (ESE), which contains the nucelotide cytosine (C) at position six in exon seven (Ex7+6), is recognised by splicing factor 2 or
alternative splicing factor (SF2/ASF), which interacts (thick black arrow) with the U2 class of small nuclear ribonuclear protein (U2 snRNP) to remove intron six. Other
splicing factors (eg, Tra2) determine splicing through interactions (thin black arrow) with ESE elements found centrally within exon seven. Serine and arginine
(SR)-rich proteins might also exert a positive splicing effect. In SMN2, the ribonucleotide uridine (transcribed from thymidine) at Ex7+6 favours exon seven exclusion
by binding to heterogeneous nuclear ribonuclear protein (hnRNP) A1, a negative splicing factor. Moreover, SF2/ASF no longer recognises this sequence motif.
Binding of hnRNP A1 is also believed to prohibit U2 snRNP binding to the branch point, which results in about 90% of SMN2 final mRNA transcripts with no exon
seven. The positive splicing factors downstream (thin black arrow) are functioning and could account for exon seven inclusion in about 10% of SMN2 transcripts.
function of an exonic splicing enhancer sequence, which exclusion of exon seven by binding heterogeneous
helps to promote normal splicing needed for production nuclear ribonuclear protein (hnRNP) A1 (a known
of intact SMN protein. This putative exonic splice splicing repressor protein). Kashima and others32 showed
enhancer sequence motif scored high for being dependent that C-to-T transition in SMN2 increases hnRNP A1
on splicing factor 2 (SF2), a positive splicing factor that is binding. The recruitment of hnRNP A1 might sterically
also known as the alternative splicing factor (ASF). The prevent the formation or stabilisation, or both, of the
SF2/ASF interacts with the U2 class of small nuclear snRNP complex needed for proper splicing. Disruption
ribonuclear protein (U2 snRNP) and its auxiliary factor of this hnRNP A1-dependent ESS with mutations or RNA
(U2AF) at the branch point within intron six to help with interference (RNAi) techniques restored SMN2 splicing
removal of the intron and successful pre-mRNA splicing to include exon seven but had no effect on SMN1.31 This
during SMN1 transcription (figure 3). Reconstruction of restoration of exon seven inclusion in SMN2 is specific
this SF2/ASF-dependent exonic splice enhancer, by to SMN.32 An A-to-G transition in intron seven (In7+100)
reverting the thymidine on Ex7+6 in SMN2 gene to in SMN2 might create a second hnRNP A1 binding site
cytosine, restored SMN2 splicing to include exon to assist further in the exclusion of exon seven.33 In an
seven.30 effort to combine both theories, Cartegni and colleagues34
Kashima and Manley31 challenged the exonic splice showed that hnRNP A1 might antagonise the SF2/ASF
enhancer hypothesis, and argued that the C-to-T exonic splice enhancer effect to promote exclusion of
nucleotide transition in exon seven creates an exonic exon seven, especially in SMN2, where this exonic splice
splice suppressor (ESS) in SMN2, which favours enhancer is inactivated. However, the exact mechanism
2122 www.thelancet.com Vol 371 June 21, 2008
Seminar
surrounding exon seven inclusion or exclusion during concentration of SMN protein resulted in low titres of
pre-mRNA splicing remains debatable. β-actin mRNA and protein in axons and growth cones,53
SMN protein is expressed in the cytoplasm and nucleus suggesting that SMN protein might be involved in
in all somatic tissues with particularly high amounts in transportation of ribonucleoprotein complexes containing
motor neurons of the spinal cord.35,36 Within the nucleus, β-actin. Deficiency of β-actin results in axonal outgrowth
this protein localises to several small (0·1–2·0 μm and pathfinding defects in two neuronal culture models54
diameter), punctate structures associated with coiled and in a zebrafish model of spinal muscular atrophy.55
(Cajal) bodies named gems (gemini of coiled bodies).37,38 Profilin, an actin-binding protein, regulates actin
Although the exact cellular function of gems remains dynamics through sequestration and release of actin
unknown, cells from patients with spinal muscular monomers. SMN protein colocalises with profilin IIa
atrophy contain substantially fewer gems than do those (the predominant isoform in neuronal tissue56) as distinct
in gene carriers and controls without the disease.36,39 granules in neurite-like extensions and growth cones.57
In eukaryotic cells, the spliceosome is the cellular Mutant SMN protein fails to interact with profilin IIa,
organelle that executes post-transcriptional pre-mRNA and knocking down profilin IIa alone inhibits neurite
splicing. Uridine-rich snRNPs (U snRNPs), the principal outgrowth.57 Setola and colleagues58 reported the
components of spliceosomes, recognise highly conserved identification of a novel SMN isoform named axonal-SMN
sequences and ligate exons. SMN protein, part of a highly that is preferentially transcribed from SMN1 and includes
stable complex with at least eight other proteins,40 is intron three. The repetitive polyadenine tract of this
necessary and sufficient for proper assembly of Smith intron was unexpectedly resistant to mutations, which
class core proteins in the U snRNP.41,42 A point mutation would alter the C-terminal protein structure, suggesting
(E134K) in the SMN tudor domain, the region that that this sequence might be needed for the structure or
mediates Smith class protein assembly,43 can cause the function, or both, of axonal-SMN.59 The axonal-SMN
disease by affecting the charge distribution in the protein was not detected in the glia or grey matter outside
SMN-Smith class binding site.44 SnRNP assembly activity Rexed’s lamina IX (motor neuron area) in the spinal
is high during embryonic and postnatal development but cord.58 Several groups have made disease models in
decreases after myogenic and neuronal differentiation. Drosophila melanogaster that show neuronal and muscular
Transcriptional networks involving many genes that defects.60,61 These models have led to postulation of roles
regulate cellular dynamics (eg, cell-cycle progression) are for SMN protein at the neuromuscular junction61 and in
highly active during this time.45,46 This RNA transcriptional skeletal muscle.60 These data suggest that SMN protein
burden might need high amounts of snRNPs to might sustain the survival of motor neurons by allowing
accomplish processing of RNA. Inhibition of snRNP normal axonal transport and maintaining the integrity of
biogenesis by defective SMN protein might cause motor neuromuscular junctions. Low concentrations of SMN
neuron degeneration seen in patients with spinal protein might be specifically detrimental to motor
muscular atrophy. Winkler and co-workers47 noted that neurons because of the long axons and their unique
impaired U snRNP biosynthesis in both frog (Xenopus interactions with skeletal muscles.
laevis) and zebrafish (Danio rerio) embryonic models The mouse Smn gene, located on chromosome 13,
induced degeneration of motor neurons similar to that contains nine exons like the human SMN gene and is
seen in spinal muscular atrophy despite rather normal 83% homologous to the SMN1 open reading frame.62,63
development. Gabanella and others48 showed that No alternative splicing isoform exists since only a
Smn–/–;SMN2+/+ mice have an eight-fold decrease in SMN full-length 1·4 kb transcript has been noted. Creating a
protein and a ten-fold decrease in snRNP assembly mouse with spinal muscular atrophy was challenging
activity in the brain and spinal cord compared with because homozygous deletion of murine Smn (Smn–/–) is
normal (Smn+/+;SMN2+/+) mice. lethal at the embryonic stage,20 whereas heterozygous
Although SMN protein is expressed in all somatic cells, Smn+/– mice are phenotypically normal. These early
why motor neurons of the spinal cord are specifically failures suggested that more innovative approaches were
vulnerable in spinal muscular atrophy is puzzling. needed to establish a mouse model of the disease.
Studies suggest ´ that SMN protein might have an In the past 7 years, several different models of spinal
important part in certain cellular functions that are muscular atrophy have been developed in mice.64
unique to motor neurons. Investigators noted that this Taiwanese scientists used the observation that patients
protein was localised in ribonucleoprotein granules in with this disease always have at least one copy of SMN2
the neurites and growth cones of primary motor neurons, and created a transgenic (Tg) mouse with human
and in motor neurons derived from embryonic stem SMN2.65 Smn+/– mice were crossed with Smn+/–;TgSMN2
cells.49,50 Live-cell fluorescence microscopy was used to mice to produce Smn–/–;TgSMN2, with clinically and
observe active bidirectional transport of these granules to histopathologically similar spinal muscular atrophy to
neuronal processes and growth cones.50 SMN protein that in people. Similar strategies were used by other
also interacts with hnRNP R and Q.51,52 hnRNP R interacts groups to create model mice that mimicked human
with the 3´ untranslated region of β-actin mRNA.51 Low spinal muscular atrophy with varying severities in
www.thelancet.com Vol 371 June 21, 2008 2123
Seminar
clinical symptoms and histopathology.66–68 Smn–/–;TgSMN2 effect. Butchback and colleagues74 created an
produced in different laboratories64,67 have varying Smn–/–;SMN2+/+;SMNΔ7+/+ mouse whose phenotype is
phenotypes, presumably because of different genetic slightly less severe (lifespan 14–15 days).64,67
backgrounds, different allelic knockouts, or different
sizes of transgene regions. By purifying the genetic Clinical implications of cloning SMN genes
background to standardise the number of SMN2 copies, Identification of the SMN genes not only enabled further
a mouse with type III spinal muscular atrophy with no research into the molecular pathogenesis of spinal
phenotypic heterogeneity was created.69 Tissue-specific muscular atrophy, but also allowed rapid and sensitive
knockdown of SMN protein has also been accomplished diagnosis. Figure 4 shows the algorithm for diagnosis of
in neurons70 or muscle,71,72 by flanking Smn with loxP the disease by molecular genetic methods. Patients with
sequences (floxed) and expressing Cre recombinase a clinical presentation resembling spinal muscular
under a neuronal-specific or muscle-specific promoter. atrophy should be tested for homozygous deletion of the
Expression of Cre recombinase results in efficient SMN1 gene, which is 95% sensitive and about 100%
deletion of floxed genes by homologous recombination. specific.75,76 Two diagnostic tests exist: a combined PCR
Constitutive overexpresssion of SMN protein has also with restriction enzyme digestion assay77,30 and an allele-
been accomplished in neuronal and skeletal muscle specific PCR assay of the SMN1 and SMN2 PCR
tissue.73 Gavrilina and others73 showed that neuronal products.78,79 The first method uses differential sus-
overexpression significantly prolonged Smn–/–;SMN2+/+ ceptibility of the SMN1 and SMN2 PCR products to
mice, but overexpression in skeletal muscle had no endonuclease digestion. The allele-specific PCR assay
SMA with typical or
atypical clincial features
SMN1 gene deletion test Homozygous SMN1 5q SMA diagnosis
deletion detected confirmed
Homozygous SMN1
deletion not detected
Repeat clinical examination,
EMG, NCS, and CK
Demyelinating or axonal Atypical SMA features, Diffuse weakness, Proximal > distal weakness,
neuropathy, NMJ disorder, neurogenic electromyography, normal electromyography, neurogenic electromyography,
myopathy, muscular dystrophy normal CK normal CK, normal NCS normal CK
Consider muscle or nerve biopsy; Consider other motor neuron Consider brain or spinal cord SMN1 gene copy count One SMN1 copy
genetic tests for myopathies, disorders (SMARD, X-linked MRI; metabolic screens
muscular dystrophies, and SMA, distal SMA, ALS)
neuropathies
Two SMN1 copies SMN1 gene sequencing SMN1 mutation found
No SMN1 mutation found 5q SMA diagnosis
confirmed
SMN-related SMA
diagnosis remains
unconfirmed
Figure 4: Diagnostic algorithm for spinal muscular atrophy
Any patient presenting with clinical symptoms resembling spinal muscular atrophy (SMA) should be tested for homozygous deletion of SMN1, which would confirm the diagnosis of SMN-associated
SMA (5q SMA). A negative SMN1 test should be followed with a repeat clinical examination for atypical clinical features (eg, contractures, eventration of hemidiaphragms, congenital absence of
muscles, pes equines deformity) and laboratory testing for creatinine phosphokinase (CK) and electrophysiological studies such as electromyography (EMG) and a nerve conduction study (NCS). If
lower motor neuron disease is suggested by EMG, SMN1 copy number will establish if SMN1 sequencing is indicated to identify intragenic mutations in patients with a single SMN1 copy. When two
SMN1 copies are detected, then investigation should be directed towards other motor neuron diseases by further diagnostic work-up such as muscle or nerve biopsy, imaging studies, metabolic
screens, and genetic testing. ALS=amyotrophic lateral sclerosis. NMJ=neuromuscular junction. SMARD=SMA with respiratory distress. Red boxes=diagnostic assessments. Blue boxes=physical and
laboratory findings. Green boxes=final diagnoses.
2124 www.thelancet.com Vol 371 June 21, 2008
Seminar
uses the nucleotide variation in exon seven and who do not want to wait for the decision until early
specifically designed PCR primers to quantitatively pregnancy, in-vitro fertilisation allows several blastomeres
amplify SMN1 or SMN2. Both tests confirm a diagnosis to be tested for SMN1 deletion after growth in culture.
of 5q spinal muscular atrophy, but the second one After the allele-specific PCR assay of a single cell is done,
provides a quantitative measure of SMN1 copy number. unaffected embryos can be implanted.94–96 This procedure
In cases in which a single copy of SMN1 is detected, requires high technical precision and is available only at
further investigation into SMN1 intragenic mutations by select centres.
gene sequencing is warranted. When two SMN1 copies Treatments to prevent motor neuron death will
are detected, further diagnostic studies should investigate presumably have the greatest effect before onset of disease.
other motor neuron, brain, spinal cord, and The challenge, however, is the identification of these
neuromuscular junction diseases (figure 4). patients before they are symptomatic. Carrier parents are
The phenotypic heterogeneity of spinal muscular asymptomatic, and most children with spinal muscular
atrophy is striking considering that nearly all patients atrophy appear normal for the first few months of life.
have a defect in the same SMN1 gene. After initial Newborn screening could address this challenge, but it has
hypotheses that the disease phenotype correlates with not become standard practice because without effective
the size of the genomic deletion,80 a thorough search of treatment this practice is not justified. A study assessed
this unstable genetic region revealed multiple SMN2 the effectiveness of genetic screening for SMN1 in neonates
copies in the human genome,81,82 possibly caused by gene with real-time quantitative PCR on 153 dried blood spots
duplication or conversion events, or both.16,83 Investigation on filter paper.97 With 100% specificity and sensitivity, this
with Smn–/– mice showed that adding a human SMN2 study provided evidence of the feasibility for large-scale
transgene could rescue pups from early embryonic newborn screening. However, social, ethical, and political
death,20 and eight copies of human SMN2 could produce implications, as well as implementation of procedures and
a phenotypically normal mouse.67 Other studies in man treatment protocols, will need to be sufficiently addressed
also showed that a higher number of SMN2 copies before screening of newborn babies can be advocated.
correlates with milder phenotypes.81 However, variations
in phenotypes are common with any number of SMN2 Treatment
copies. Therefore, prediction of the spinal muscular One substantial implication of uncovering the molecular
atrophy phenotype with SMN2 copy number is not genetic basis of spinal muscular atrophy is in the devel-
recommended. Some evidence suggests that other opment of candidate therapeutics. The unique genomic
phenotype-modifier genes—eg, GTF2H2 (BTF2P44), organisation of the SMN genes allows a novel therapeutic
SERF1A (H4F5)—might exist.84,85 Evidence also suggests approach: to promote SMN2, existing in all patients, to
that higher amount of SMN protein is observed in function like the missing SMN1 gene. This idea prompted
patients with milder form of spinal muscular atrophy.86 investigations into increasing inclusion of exon seven in
High homology between SMN1 and SMN2 complicated SMN2 mRNA transcripts,39,98–101 upregulation of SMN2
carrier detection with PCR methods, because transcription by promoter activation,102–104 modulation of
amplification of an SMN1 fragment frequently amplifies SMN protein translation,105–107 and prevention of SMN
SMN2 as well. Initial carrier detection methods used protein degradation.28,108
high sensitivity densitometry of PCR products to identify Serine–arginine-rich proteins are splicing factors that
copy number.82 Present methods use quantitative interact with exonic splice enhancers and affect mRNA
real-time PCR allowing measurement of both SMN1 and splicing. Htra2-β1, a serine–arginine-rich-like splicing
SMN2 copy number separately.78,80,87,88 However, the SMN1 factor, was found to stimulate full-length SMN2
copy number test might rarely and inaccurately determine expression109 by interacting with exonic splice enhancers,
the carrier status, because about 3·2% of the general especially elements that contain AG-rich motifs such as
population has both SMN1 copies on one chromosome positive splicing enhancer 2.26 Abrogation of such motifs
and none on the other (denoted 2+0).89,90 These people are results in skipping of exon seven.26 Little is known about
carriers because one of their chromosomes is missing a the importance of Htra2-β1, and SMN genes are the only
normal SMN1 allele. Another 1·7% have intragenic known human genes whose splicing is specifically
mutations that are not detectable by these methods of regulated by Htra2-β1; other human isoforms (eg,
SMN1 dosage analysis.80 SMN1 gene sequencing can Htra2-β3 and Htra2-α) did not stimulate proper SMN2
detect intragenic mutations, and 2+0 carriers can be splicing.109 The use of pharmacological compounds to
detected by karyotyping with fluorescence in-situ upregulate Htra2-β1 might prove to be a valuable strategy
hybridisation. to modulate SMN2 splicing.
Chorionic villi sampling and amniocentesis can One of the first candidate treatments for spinal
effectively find out the fetal genotype.91,92 Additionally, muscular atrophy was aclarubicin (aclacinomycin A), an
circulating fetal cells in maternal blood can be isolated by anthracycline and topoisomerase II inhibitor used in
isolation by size of epithelial tumour or trophoblastic patients with solid tumours or leukaemia. Aclarubicin
cells (ISET) for PCR-based genetic analysis.93 For those was discovered by screening a 960-member combinatorial
www.thelancet.com Vol 371 June 21, 2008 2125
Seminar
chemistry library with a semiquantitative assay based on 550 000 compounds in a β-lactamase assay designed to
RT-PCR to detect inclusion of exon seven.99 Aclarubicin’s detect activation of the SMN2 promoter. They discovered
toxicity prohibited further development for clinical use to five classes of compounds that altered SMN2 transcript.
treat patients with spinal muscular atrophy. Nevertheless, Four of these increased the full-length to truncated SMN2
it served as a proof of principle for identifying modifiers transcript ratio. Two also increased SMN protein and the
of SMN2 splicing and that high throughput screening number of nuclear gems in fibroblasts from type I
was a valid approach to identify candidates. patients. Further medicinal chemistry and structure-
With histone acetylation, the surrounding DNA relaxes activity relationship studies will assess the promise of
and is accessible to transcriptional machinery. Inhibition these molecules in clinical use.
of histone deacetylation might enhance expression in Grzeschik and colleagues39 noted that hydroxyurea, a
about 2% of human genes.110 Sodium butyrate was the ribonucleotide reductase inhibitor, increased the full length
first histone deacetylase inhibitor found to enhance to truncated SMN mRNA ratio. The total amount of SMN
SMN2 expression and increase exon seven inclusion in mRNA in the lymphoblastoid cell lines from patients with
lymphoid cell lines derived from patients with spinal spinal muscular atrophy was not changed after hydroxyurea
muscular atrophy. This treatment also increased treatment, but the increase in full-length SMN mRNA was
expression of SMN protein in the spinal cord and motor coupled by a decrease in truncated SMN mRNA. This
neurons of SMA-like mice developed by the Taiwanese finding shows that hydroxyurea increased the full-length
researchers described previously.99 Not surprisingly, the SMN mRNA by promoting exon seven inclusion rather
expression of other genes (eg, globin) was also increased. than activating the SMN2 promoter.39 Furthermore,
However, the short half-life, about 6 min in human hydroxyurea treatment, in vitro, increased SMN protein
serum, prohibited sodium butyrate from substantial and the number of nuclear gems. Hydroxyurea also has a
clinical development. Encouraged by the effect of sodium good safety profile, easy oral administration, and high
butyrate, the search began for other putative histone bioavailability. Riluzole is a neuroprotective agent known
deacetylase inhibitors. In 2003, two groups reported that to promote neuronal survival through glutaminergic
valproic acid, a histone deacetylase inhibitor approved for antagonism.116 Haddad and colleagues117 treated special
epilepsy treatment, increased full-length SMN mRNA. spinal muscular atrophy mice with Smn exon seven deleted
Valproic acid activated the SMN2 promoter and induced only in neurons and found that riluzole extended lifespan
expression of positive splicing factors, SF2/ASF and but did not significantly improve function. It prevented
Htra2-β1.100,101 In 2004, Andreassi and colleagues103 aberrant cytoskeletal organisation at the motor terminals
reported that a sodium butyrate derivative, phenylbutyrate, but did not prevent loss of proximal axons.117 After screening
increased full-length transcript production from SMN2, about 47 000 compounds, Lunn and others107 discovered
SMN protein, and the number of nuclear gems. that indoprofen, a non-specific cyclo-oxygenase inhibitor
Phenylbutyrate is still used to treat children with urea- and a non-steroidal anti-inflammatory drug, increased
cycle defects and has a slightly longer half-life (0·8–1·0 h SMN protein in fibroblasts of patients with type I spinal
in human serum) than has sodium butyrate.111 muscular atrophy. It also increased the number of nuclear
Further investigations were directed towards more gems and caused a trend towards increased viability in the
potent and specific histone deacetylase inhibitors. Smn–/–;TgSMN2 spinal muscular atrophy mouse model.107
Suberoylanilide hydroxamic acid, a potent histone No other non-steroidal anti-inflammatory drugs that were
deacetylase inhibitor under clinical investigation for tested showed a similar effect, suggesting that indoprofen’s
cancer therapy,112 has good oral bioavailability and is effect is independent of cyclo-oxygenase inhibition. A
non-toxic, despite complete inhibition.113 At low previously approved drug in the UK, indoprofen, was
micromolar concentration, suberoylanilide hydroxamic removed from the market because of high frequency of
acid activated the SMN2 gene in fibroblasts from patients gastrointestinal bleeding secondary to cyclo-oxygenase
and increased SMN protein in rat motor neuron cultures, inhibition. Other compounds—eg, a β-adrenergic
and in both human and rat hippocampal brain slice agonist,118 an Na+/H+ exchanger inhibitor,119 and polyphenol
cultures.113 plant compounds120—have been reported to promote exon
Another potent hydroxamic acid, trichostatin A, was seven inclusion and to increase SMN protein concentration
originally isolated in 1976 as an antifungal drug,114 but it in fibroblasts derived from patients with spinal muscular
has not been in clinical use because of the high cost of atrophy.
isolation and purification. Treatment with trichostatin A Translation of SMN1 mRNA ends at a stop codon at the
in normal and SMNΔ7 mouse models specifically end of exon seven with no translation of exon eight. The
increased properly spliced SMN2 transcript, snRNP 16 aminoacids absent in SMNΔ7 protein, although not a
assembly, and SMN protein in neural and muscle tissue.115 direct mediator of SMN’s numerous activities, are
Even after the onset of disease, trichostatin A treatment believed to be required for oligomerisation of SMN
was able to extend survival and increase the size of protein.121,122 Heterologous sequences are able to
myofibres and spinal cord motor neurons in the SMNΔ7 substitute123 for the cytoplasmic localisation signal of
mouse model. Jarecki and colleagues104 screened SMN protein.50 Since aminoglycosides have been reported
2126 www.thelancet.com Vol 371 June 21, 2008
Seminar
to promote stop codon read-through,124 the Lorson to increased exon seven inclusion, the amount of SMN
group105,106 reported that various aminoglycosides in- protein, and the number of nuclear gems in fibroblasts
creased the number of nuclear gems and full-length from patients with spinal muscular atrophy. Some
SMN protein by preventing recognition of stop codon in bifunctional antisense oligonucleotides also have a
the SMN transcript; this read-through results in the non-complementary tail that is rich in potent exonic
addition of a non-specific tail needed for proper cyto- splice enhancer sequences to recruit trans-acting
plasmic localisation of the SMN gene and increased positive factors that might inhibit binding of negative
activity of SMNΔ7 protein. splicing proteins.126 Antisense oligonucleotides were
Another way to modulate SMN2 splicing was to use able to modulate splicing and inhibit signalling in a
exon-specific antisense oligonucleotides that were mouse study.127 Further investigations into bifunctional
complementary to regions of SMN2 exon seven.125,126 antisense oligonucleotides might increase potency but
Antisense oligonucleotides bound to pre-mRNA run the risk of being incompatible as a treatment option
inhibited binding of negative splicing factors, which led because of possible endogenous immunological
Design SMA type Sample size Primary outcome Secondary outcome Results Comments
measure measure
Acetyl-L-carnitine MC, randomisation, DB, II, III 110 Handgrip and elbow Knee flexion strength, Completed, results pending http://www.enmc.org/
PC flexion strength forced vital capacity, workshop/?id=73
quality of life
Albuterol131 SC, OL, PS II, III 13 (5 type II, Muscle strength, .. Significant increase in all No placebo control,
8 type III) forced vital capacity, outcome measures unblinded. Improvement
lean body mass could be due to normal
growth
Creatine MC, randomisation, PC II, III 55 GMFM Functional tests, Completed, results pending No effect
pulmonary function tests
Gabapentin136,140 MC, randomisation, DB, II, III 84 (40 treatment, Muscle strength Forced vital capacity, No difference between ..
PC 44 placebo) testing SMA rating scale, mini- treatment and placebo
sickness impact profile groups
MC, randomisation, II, III 120 (61 treatment, Change in maximum Forced vital capacity, Improvement in leg strength ..
treatment vs non 59 no treatment) voluntary isometric timed tests and a trend for improvement
treatment contraction in arm strength
Hydroxyurea141 SC, PI or PII, I 18 (2:1 treated to Survival MUNE, SMN mRNA and In progress ClinicalTrials.gov
randomisation, DB, PC placebo ratio) protein Identifier: NCT00083746
SC, PI or PII, II or III 24 (2:1 treated to GMFM MUNE, SMN mRNA and In progress ClinicalTrials.gov
randomisation, DB, PC placebo ratio) protein Identifier: NCT00084006
SC, PII, randomisation, II or III 60 SMN mRNA and .. In progress ClinicalTrials.gov
DB, PC protein, motor Identifier: NCT00485511
function, lung
function
SC, OL, PS II, III 33 HFMS SMN mRNA Completed ..
Phenylbutyrate134–136 MC, PI or PII, OL, PS II 29 (10 treatment, Tolerability HFMS, myometry, forced Significant increase in ..
19 no treatment) vital capacity HFMS scores
SC, OL, PS II, III 6 (4 type II, SMN mRNA level .. Increased SMN mRNA level ..
2 type III) and 3
carriers
MC, PII, randomisation, II 107 (54 treatment, HFMS Myometry, forced vital No significant improvement ..
DB, PC 53 placebo) capacity
MC, PI, II, III 30 Tolerability SMN mRNA and protein In progress ClinicalTrials.gov
OL Identifier: NCT00439569
MC, PI, I 30 Tolerability Pharmacokinetics, SMN In progress ClinicalTrials.gov
OL mRNA and protein Identifier: NCT00439218
SC, PI or PII, OL II, III 12 Safety, tolerability Overall motor function In progress ClinicalTrials.gov
Identifier: NCT00528268
SC, OL, PS I, II 20 Safety SMN mRNA and protein, Completed, results pending ··
TIMP, HFMS
Riluzole139 MC, PI, randomisation, I 10 (7 treatment, Survival .. No adverse event or serious Trial prematurely stopped
DB, PC 3 placebo) adverse event attributed to due to withdrawn
treatment financial support
MC, PI or II, OL I 40 (targeted), Survival SMN mRNA Completed, results pending ..
3 (completed)
(Continues on next page)
www.thelancet.com Vol 371 June 21, 2008 2127
Seminar
Design SMA type Sample size Primary outcome Secondary outcome Results Comments
measure measure
(Continued from previous page)
Somatotropin MC, PII, randomisation, II, III 20 Hand-held myometry Functional tests, Not yet recruiting ClinicalTrials.gov
DB, PC, crossover pulmonary function Identifier: NCT00533221
testing
Thyrotropin- SC, randomisation, DB, II, III 9 (6 treatment, Muscle strength using NCV, CMAP Improvement muscle Patient served as own
releasing hormone PC 3 placebo) hand-held strength and some CMAP/ control because of small
(protirelin)142 dynamometry NCV values sample size
Valproic acid137,138 SC, OL, PS I, II, III 21 (5 type I, SMN2 mRNA level .. SMN2 mRNA increased in ..
10 type II, 6 type III) 7 patients with SMA,
and 10 carriers unchanged or decreased in 13
SC, Retrospective, OL III, IV 7 Hand-held .. Strength improvement by ..
dynamometry 48%
SC, PII, randomisation, Type III 36 Muscle strength MUNE, lean body mass, In progress ClinicalTrials.gov
DB, PC, crossover ambulatory timed tests Identifier: NCT00481013
SC, PI or PII, OL, PS I, II, III 42 Tolerability SMN mRNA and protein, Completed ClinicalTrials.gov
(>2 years) EMG, pulmonary Identifier: NCT00374075
function testing
Valproic acid and PII, randomisation, DB, Cohort 1: 90 (in two cohorts) Safety, HFMS, SMN mRNA, MUNE, In progress ClinicalTrials.gov
L-carnitine PC, crossover sitting II and myometry CMAP, pulmonary Identifier: NCT00227266
III; cohort 2: function testing
standing II
and III
The information contained in the ongoing studies was gathered through unpublished sources, and some might not be current or complete. CMAP=compound motor unit action potentials. DB=double-blind.
GMFM=gross motor function measure. HFMS=Hammersmith functional motor score. MC=multicentre. MUNE=motor unit number estimation. NCV=nerve conduction velocities. OL=open-label.
PC=placebo-controlled. PI=phase I. PII=phase II. PIII=phase III. PS=pilot study. SC=single centre. TIMP=test of infant motor performance.
Table 2: Completed and ongoing clinical trials of various therapeutic agents for spinal muscular atrophy
reactions. Cartegni and Krainer128 developed a peptide at ten Italian centres, however, reported no significant
nucleic acid oligonucleotide mimetic that is resistant to change in functional motor scores after 13 weeks of
nucleases and does not activate ribonuclease H on treatment.135 Similarly, a placebo-controlled, randomised,
binding to the target RNA. High amounts of this nucleic double-blind trial of gabapentin in types II and III
acid mimetic increased exon seven inclusion in a patients showed no substantial difference in muscle
three-exon (exon six through eight) mini-gene splicing strength between treatment and placebo groups after
reaction in vitro. This effect was sequence specific, 12 months of treatment.136 Treatment of patients with
because a peptide nucleic acid directed against another spinal muscular atrophy and disease-gene carriers with
gene had no effect. DiMatteo and colleagues129 used valproic acid in a pilot study resulted in an increase in
single-stranded oligonucleotides to convert SMN2 into full-length SMN expression, but no clinical outcome was
SMN1 in fibroblasts of patients with spinal muscular measured.137 An open-label study of valproic acid in seven
atrophy. The conversion was confirmed by quantitative patients with mild disease (types III and IV) showed a
RT-PCR and by proper intracellular SMN protein rise in quantitative muscle strength by dynamometry
localisation. and improved subjective motor functioning after a mean
treatment duration of 8 months.138 Riluzole was tested in
Clinical trials a phase I, randomised, double-blind, placebo-controlled
Many compounds noted to increase SMN mRNA and study for patients with type I disease.139 Although this
protein expression in vitro and in animal models in the study lacked statistical power because of a small sample
past few years led to human studies of candidate size, benefit was suggested in treated patients. A
therapeutics.130 Small-scale studies in man with candidate multicentre randomised trial intended to further study
compounds that have no supporting preclinical data have the effect of riluzole in type I has been completed;
met little success.131,132 Several clinical trials based on however, the result is pending. Several randomised trials
strong pre-clinical data are in progress. Early pilot studies are in progress, including a multicentre trial with
of phenylbutyrate in patients with types II and III spinal combined l-carnitine and valproic acid; two double-blind,
muscular atrophy showed increases in full-length SMN placebo-controlled trials with hydroxyurea in type I and
expression and in motor function as measured by in types II or III patients; and a multicentre trial
myometry and the Hammersmith functional motor assessing the efficacy of somatotropin in type II and III
scale.133,134 A randomised, double-blind, placebo-controlled patients. Table 2 lists the clinical trials completed and
trial of phenylbutyrate in 107 patients with type II disease in progress.
2128 www.thelancet.com Vol 371 June 21, 2008
Seminar
Conclusion noted in ventral roots.151 Moreover, these motor neurons
Future advances in high-throughput screening technol- formed neuromuscular junctions and induced contraction
ogies, computation, and organic synthesis143 will allow in vitro with cultured myoblasts. Future research would
development of specific, non-toxic, potent small molecules need to aim at delivering these stem cells derived from
to modulate splicing and expression of SMN2. motor neurons to the spinal cord, countering host versus
Government-backed programmes, such as the SMA graft immunological responses, and guiding axonal For more on the SMA project
Project, will accelerate these goals. Additionally, modulation outgrowth to reach the target muscles. see http://www.smaproject.org
of non-SMN targets (eg, Gemin2 and Gemin6) can affect As clinical trials have progressed, several challenges
SMN protein activity.144 Promising, alternative gene therapy have surfaced.151 The small number of patients for clinical
approaches to modulate pathogenesis with viral vectors to trials will need a coordinated effort for patients to
deliver SMN1 directly,145 trans-splicing RNAs to correct participate in different trials. Criteria will need to be set up
SMN2 splicing,146 and neurotrophic agents (ie, for the selection of compounds for clinical trials. Various
cardiotrophin-1)147 should be explored. Motor neurons elements of trial design will need to be unified to allow
derived from stem cells have been established,148,149 and comparisons of results between trials. Sensitive and valid
stem cells are promising for use in assays and human outcome measures will need to be developed.130 Few
therapeutic trials. Neuronal stem cells proved to protect clinical outcome measures are available for patients with
surviving motor neurons in rats with motor neuron type I spinal muscular atrophy other than survival.
injury.150 In an exciting report, embryonic stem cells derived Although myometry using hand-held dynamometers152
from motor neurons survived in spinal cords of rats with and quantitative muscle testing153,154 are reliable muscle
motor neuron injury, and axons from these cells were strength measures in types II and III patients, these are
not useful for type I patients. Functional motor scales,
Panel: Databases and websites such as the Hammersmith functional motor scale and
gross motor functional measurement, are also restricted
Databases to types II and III patients.155 Measuring the function of
• Online Mendelian Inheritance in Man (NCBI/OMIM): other organ systems, such as pulmonary function and
http://www.ncbi.nlm.nih.gov/omim/ ability to swallow and speak, might be sensitive outcome
• PubMed: http://www.pubmed.com measures, especially in younger patient groups.
Clinical trial networks in the USA Additionally, biomarkers, such as SMN mRNA and protein
• American SMA Randomised Trial (AmSMART) Group: and gem count, might be sensitive outcome measures for
http://www.amsmart.org clinical trials. The amounts of full-length SMN transcripts
• Pediatric Neuromuscular Clinical Research Network and SMN protein are well correlated with disease severity.
(PNCR): http://www.urmc.edu/sma As such, several groups have developed promising assays
• Project Cure SMA: http://www.projectcuresma.org to monitor changes in SMN mRNA and protein in human
blood.156,157SMN mRNA seems to be the most amenable to
Clinical trial networks in Europe large multicentre trials.158
• European Neuromuscular Centre (ENMC): The clinical implications of elucidating the molecular
http://www.enmc.org genetic basis of spinal muscular atrophy and advances in
• TREAT-NMD: http://www.treat-nmd.eu medical technology have changed the clinical care of
Clinical trial registry in the USA patients. However, these patients still receive highly
• ClinicalTrials.gov: http://www.clinicaltrials.gov diverse care for several reasons such as large clinical
phenotypic variation, multiorgan system involvement,
Patient and family advocacy groups geographical variation in availability of medical expertise,
• The Benjamin Foundation: different values of clinicians and families, and variations
http://www.thebenjaminfoundation.org in financial resources. Differences in clinical care also
• Families of SMA: http://www.fsma.org hinder valid measures of clinical outcomes in clinical
• Fight SMA/Andrew’s Buddies: http://www.fightsma.org trials. The International Standard of Care Committee for
• Hope and Light Foundation: http://www.hopeandlight.org spinal muscular atrophy was established in 2005 with a
• Miracle for Madison and Friends: mission to set a standard of care. The committee consists
http://www.miracleformadison.org of 12 core members and four working groups: diagnostics
• Payton’s Pals: http://www.paytonspals.com or new interventions, pulmonary, gastroenterology or
• SMA Angels Charity: http://www.smaangels.org nutrition, and orthopaedics or rehabilitation. Each
• SMA Foundation: http://www.smafoundation.org working group contains eight to ten experts in that
• SMA Support: http://www.smasupport.com specialty. The groups identified few data that were
Patient registry available to establish evidence-based practice guidelines
• International SMA Patient Registry: http://www.iupui. for the disease. Hence, a Delphi survey was used to
edu/~medgen/hereditary/sma.html achieve consensus among more than 60 experts from
12 countries. The committee held two meetings to draft a
www.thelancet.com Vol 371 June 21, 2008 2129
Seminar
consensus statement for the standard of care for patients.7 16 Burghes AH. When is a deletion not a deletion? When it is
converted. Am J Hum Genet 1997; 61: 9–15.
As a result, we hope that receiving consistent medical
17 McLean MD, Roy N, MacKenzie AE, et al. Two 5q13 simple tandem
care will become easier for patients and that valid clinical repeat loci are in linkage disequilibrium with type 1 spinal muscular
outcomes can be measured during clinical trials. atrophy. Hum Mol Genet 1994; 3: 1951–56.
Although major challenges remain towards developing 18 Lefebvre S, Burglen L, Reboullet S, et al. Identification and
characterization of a spinal muscular atrophy-determining gene.
a therapeutic agent for patients with spinal muscular Cell 1995; 80: 155–65.
atrophy, our understanding of the disease biology, 19 Burglen L, Lefebvre S, Clermont O, et al. Structure and
including several animal models, has grown exponentially organization of the human survival motor neurone (SMN) gene.
Genomics 1996; 32: 479–82.
since identification of SMN1 in 1995. Clinical trials have
20 Schrank B, Gotz R, Gunnersen JM, et al. Inactivation of the survival
begun and have uncovered areas to improve the design motor neuron gene, a candidate gene for human spinal muscular
of human studies and patient care (panel). Further atrophy, leads to massive cell death in early mouse embryos.
Proc Natl Acad Sci USA 1997; 94: 9920–25.
research and development, coupled with a broadly
21 Paushkin S, Charroux B, Abel L, Perkinson RA, Pellizzoni L,
accepted and applied standard of care for these patients, Dreyfuss G. The survival motor neuron protein of
will help us manage, treat, and eventually cure this Schizosacharomyces pombe: conservation of survival motor neuron
interaction domains in divergent organisms. J Biol Chem 2000;
devastating neurodegenerative disease. 275: 23841–46.
Conflict of interest statement 22 Miguel-Aliaga I, Culetto E, Walker DS, Baylis HA, Sattelle DB,
We declare that we have no conflict of interest. We have no relation to Davies KE. The Caenorhabditis elegans orthologue of the human
any patent application or intellectual property for any spinal muscular gene responsible for spinal muscular atrophy is a maternal product
atrophy therapeutic candidate. critical for germline maturation and embryonic viability.
Hum Mol Genet 1999; 8: 2133–43.
Acknowledgments 23 Rochette CF, Gilbert N, Simard LR. SMN gene duplication and the
We thank Rishi Bhatnagar for medical illustration and Hannes Vogel for emergence of the SMN2 gene occurred in distinct hominids: SMN2
providing histopathological slides. MRL is supported by the Molecular is unique to Homo sapiens. Hum Genet 2001; 108: 255–66.
Basis of Medicine scholarly concentration at Stanford University School 24 Hahnen E, Forkert R, Marke C, et al. Molecular analysis of
of Medicine. candidate genes on chromosome 5q13 in autosomal recessive
spinal muscular atrophy: evidence of homozygous deletions of
References
the SMN gene in unaffected individuals. Hum Mol Genet 1995;
1 Pearn J. Classification of spinal muscular atrophies. Lancet 1980;
4: 1927–33.
1: 919–22.
25 Lorson CL, Androphy EJ. An exonic enhancer is required for
2 Ogino S, Leonard DG, Rennert H, Ewens WJ, Wilson RB. Genetic
inclusion of an essential exon in the SMA-determining gene SMN.
risk assessment in carrier testing for spinal muscular atrophy.
Human Molecul Genet 2000; 9: 259–65.
Am J Med Genet 2002; 110: 301–07.
26 Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide
3 Buchthal F, Olsen PZ. Electromyography and muscle biopsy in
in the SMN gene regulates splicing and is responsible for spinal
infantile spinal muscular atrophy. Brain 1970; 93: 15–30.
muscular atrophy. Proc Natl Acad Sci USA 1999; 96: 6307–11.
4 Emery AE, Hausmanowa-Petrusewicz I, Davie AM, Holloway S,
27 Vitte J, Fassier C, Tiziano FD, et al. Refined characterization of the
Skinner R, Borkowska J. International collaborative study of the
expression and stability of the SMN gene products. Am J Pathol
spinal muscular atrophies. Part 1. Analysis of clinical and laboratory
2007; 171: 1269–80.
data. J Neurol Sci 1976; 29: 83–94.
28 Chang HC, Hung WC, Chuang YJ, Jong YJ. Degradation of survival
5 Munsat TL. International SMA Collaboration. Neuromuscul Disord
motor neuron (SMN) protein is mediated via the ubiquitin/
1991; 1: 81.
proteasome pathway. Neurochem Int 2004; 45: 1107–12.
6 Russman BS. Spinal muscular atrophy: clinical classification and
29 Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH. Differential
disease heterogeneity. J Child Neurol 2007; 22: 946–51.
SMN2 expression associated with SMA severity. Nat Genet 1998;
7 Wang CH, Finkel RS, Bertini ES, et al. Consensus statement for 20: 230–31.
standard of care in spinal muscular atrophy. J Child Neurol 2007;
30 Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent
22: 1027–49.
exonic splicing enhancer in SMN2 causes spinal muscular atrophy
8 Markowitz JA, Tinkle MB, Fischbeck KH. Spinal muscular in the absence of SMN1. Nat Genet 2002; 30: 377–84.
atrophy in the neonate. J Obstet Gynecol Neonatal Nurs 2004;
31 Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits
33: 12–20.
splicing in spinal muscular atrophy. Nat Genet 2003; 34: 460–63.
9 Brzustowicz LM, Lehner T, Castilla LH, et al. Genetic mapping of
32 Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with
chronic childhood-onset spinal muscular atrophy to chromosome
specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet
5q11.2-13.3. Nature 1990; 344: 540–41.
2007; 16: 3149–59.
10 Melki J, Abdelhak S, Sheth P, et al. Gene for chronic proximal
33 Kashima T, Rao N, Manley JL. An intronic element contributes to
spinal muscular atrophies maps to chromosome 5q. Nature 1990;
splicing repression in spinal muscular atrophy.
344: 767–78.
Proc Natl Acad Sci USA 2007; 104: 3426–31.
11 Brzustowicz LM, Kleyn PW, Boyce FM, et al. Fine-mapping of the
34 Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR.
spinal muscular atrophy locus to a region flanked by MAP1B and
Determinants of exon 7 splicing in the spinal muscular atrophy
D5S6. Genomics 1992; 13: 991–98.
genes, SMN1 and SMN2. Am J Hum Genet 2006; 78: 63–77.
12 Wang CH, Kleyn PW, Vitale E, et al. Refinement of the spinal
35 Battaglia G, Princivalle A, Forti F, Lizier C, Zeviani M. Expression
muscular atrophy locus by genetic and physical mapping.
of the SMN gene, the spinal muscular atrophy determining gene, in
Am J Hum Genet 1995; 56: 202–09.
the mammalian central nervous system. Hum Mol Genet 1997;
13 Clermont O, Burlet P, Burglen L, et al. Use of genetic and physical 6: 1961–71.
mapping to locate the spinal muscular atrophy locus between two new
36 Coovert DD, Le TT, McAndrew PE, et al. The survival motor neuron
highly polymorphic DNA markers. Am J Hum Genet 1994; 54: 687–94.
protein in spinal muscular atrophy. Hum Mol Genet 1997; 6: 1205–14.
14 Schmutz J, Martin J, Terry A, et al. The DNA sequence and
37 Liu Q, Dreyfuss G. A novel nuclear structure containing the
comparative analysis of human chromosome 5. Nature 2004;
survival of motor neurons protein. Embo J 1996; 15: 3555–65.
431: 268–74.
38 Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT,
15 Melki J, Lefebvre S, Burglen L, et al. De novo and inherited
Carmo-Fonseca M. The spinal muscular atrophy disease gene
deletions of the 5q13 region in spinal muscular atrophies. Science
product, SMN: A link between snRNP biogenesis and the Cajal
1994; 264: 1474–77.
(coiled) body. J Cell Biol 1999; 147: 715–28.
2130 www.thelancet.com Vol 371 June 21, 2008
Seminar
39 Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH. 60 Gunadi, Sasongko TH, Yusoff S, et al. Hypomutability at the
Hydroxyurea enhances SMN2 gene expression in spinal muscular polyadenine tract in SMN intron 3 shows the invariability of the
atrophy cells. Ann Neurol 2005; 58: 194–202. a-SMN protein structure. Ann Hum Genet 2008; 72: 288–91.
40 Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, 61 Chan YB, Miguel-Aliaga I, Franks C, et al. Neuromuscular defects in a
Fischer U. A comprehensive interaction map of the human survival Drosophila survival motor neuron gene mutant. Hum Mol Genet 2003;
of motor neuron (SMN) complex. J Biol Chem 2007; 282: 5825–33. 12: 1367–76.
41 Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN 62 Bergin A, Kim G, Price DL, Sisodia SS, Lee MK, Rabin BA.
complex in the specificity of snRNP assembly. Science 2002; Identification and characterization of a mouse homologue of the
298: 1775–79. spinal muscular atrophy-determining gene, survival motor neuron.
42 Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A Gene 1997; 204: 47–53.
multiprotein complex mediates the ATP-dependent assembly of 63 DiDonato CJ, Chen XN, Noya D, Korenberg JR, Nadeau JH,
spliceosomal U snRNPs. Nat Cell Biol 2001; 3: 945–49. Simard LR. Cloning, characterization, and copy number of the
43 Buhler D, Raker V, Luhrmann R, Fischer U. Essential role for the murine survival motor neuron gene: homolog of the spinal
tudor domain of SMN in spliceosomal U snRNP assembly: muscular atrophy-determining gene. Gen Res 1997; 7: 339–52.
implications for spinal muscular atrophy. Hum Mol Genet 1999; 64 Schmid A, DiDonato CJ. Animal models of spinal muscular
8: 2351–57. atrophy. J child Neurol 2007; 22: 1004–12.
44 Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. 65 Hsieh-Li HM, Chang JG, Jong YJ, et al. A mouse model for spinal
SMN tudor domain structure and its interaction with the Sm muscular atrophy. Nat Genet 2000; 24: 66–70.
proteins. Nat Struct Biol 2001; 8: 27–31. 66 Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M.
45 Cam H, Balciunaite E, Blais A, et al. A common set of gene Reduced survival motor neuron (Smn) gene dose in mice leads to
regulatory networks links metabolism and growth inhibition. motor neuron degeneration: an animal model for spinal muscular
Mol Cell 2004; 16: 399–411. atrophy type III. Hum Mol Genet 2000; 9: 341–46.
46 Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, 67 Monani UR, Sendtner M, Coovert DD, et al. The human centromeric
Dynlacht BD. An initial blueprint for myogenic differentiation. survival motor neuron gene (SMN2) rescues embryonic lethality in
Genes Dev 2005; 19: 553–69. Smn(–/–) mice and results in a mouse with spinal muscular atrophy.
47 Winkler C, Eggert C, Gradl D, et al. Reduced U snRNP assembly Hum Mol Genet 2000; 9: 333–39.
causes motor axon degeneration in an animal model for spinal 68 Monani UR, Pastore MT, Gavrilina TO, et al. A transgene carrying
muscular atrophy. Genes Dev 2005; 19: 2320–30. an A2G missense mutation in the SMN gene modulates phenotypic
48 Gabanella F, Butchbach MER, Saieva L, et al. Ribonucleoprotein severity in mice with severe (type I) spinal muscular atrophy.
assembly defects correlate with spinal muscular atrophy severity J Cell Biol 2003; 160: 41–52.
and preferentially affect a subset of spliceosomal snRPs. PLoS ONE 69 Tsai LK, Tsai MS, Lin TB, Hwu WL, Li H. Establishing a
2007; 2: e921 standardized therapeutic testing protocol for spinal muscular
49 Fan L, Simard LR. Survival motor neuron (SMN) protein: role in atrophy. Neurobiol Dis 2006; 24: 286–95.
neurite outgrowth and neuromuscular maturation during neuronal 70 Frugier T, Tiziano FD, Cifuentes-Diaz C, et al. Nuclear targeting
differentiation and development. Hum Mol Genet 2002; 11: 1605–14. defect of SMN lacking the C-terminus in a mouse model of spinal
50 Zhang H, Xing L, Rossoll W, Wichterle H, Singer RH, Bassell GJ. muscular atrophy. Hum Mol Genet 2000; 9: 849–58.
Multiprotein complexes of the survival of motor neuron protein 71 Ferri A, Melki J, Kato AC. Progressive and selective degeneration of
SMN with Gemins traffic to neuronal processes and growth cones motoneurons in a mouse model of SMA. Neuroreport 2004;
of motor neurons. J Neurosci 2006; 26: 8622–32. 15: 275–80.
51 Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, 72 Cifuentes-Diaz C, Frugier T, Tiziano FD, et al. Deletion of murine
Sendtner M. Specific interaction of Smn, the spinal muscular SMN exon 7 directed to skeletal muscle leads to severe muscular
atrophy determining gene product, with hnRNP-R and gry-rbp/ dystrophy. J Cell Biol 2001; 152: 1107–14.
hnRNP-Q: a role for Smn in RNA processing in motor axons? 73 Gavrilina TO, McGovern VL, Workman E, et al. Neuronal SMN
Hum Mol Genet 2002; 11: 93–105. expression corrects spinal muscular atrophy in severe SMA mice
52 Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts while muscle specific SMN expression has no phenotypic effect.
with a novel family of hnRNP and spliceosomal proteins. Embo J Hum Mol Genet (in press).
2001; 20: 5443–52. 74 Butchback MER, Edwards JD, Burghes AHM. Abnormal motor
53 Rossoll W, Jablonka S, Andreassi C, et al. Smn, the spinal muscular phenotype in the SMNΔ7 mouse model of spinal muscular atrophy.
atrophy-determining gene product, modulates axon growth and Neurobiol Dis 2007; 27: 207–19.
localization of beta-actin mRNA in growth cones of motoneurons. 75 Lefebvre S, Burglen L, Frezal J, Munnich A, Melki J. The role of the
J Cell Biol 2003; 163: 801–12. SMN gene in proximal spinal muscular atrophy. Hum Mol Genet
54 Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. 1998; 7: 1531–36.
Active transport of the survival motor neuron protein and the role 76 Rodrigues NR, Owen N, Talbot K, Ignatius J, Dubowitz V,
of exon-7 in cytoplasmic localization. J Neurosci 2003; 23: 6627–37. Davies KE. Deletions in the survival motor neuron gene on 5q13 in
55 McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown autosomal recessive spinal muscular atrophy. Hum Mol Genet 1995;
of the survival motor neuron (Smn) protein in zebrafish causes 4: 631–34.
defects in motor axon outgrowth and pathfinding. J Cell Biol 2003; 77 van der Steege G, Grootscholten PM, van der Vlies P, et al.
162: 919–31. PCR-based DNA test to confirm clinical diagnosis of autosomal
56 Lambrechts A, Braun A, Jonckheere V, et al. Profilin II is recessive spinal muscular atrophy. Lancet 1995; 345: 985–86.
alternatively spliced, resulting in profilin isoforms that are 78 Ichikawa S, Wang CH. Differential amplification of the SMN gene
differentially expressed and have distinct biochemical properties. copies by allele-specific PCR: a new method for rapid diagnosis of
Mol Cell Biol 2000; 20: 8209–19. spinal muscular atrophy. Am J Hum Genet 1998; 63: A232.
57 Sharma A, Lambrechts A, Hao le T, et al. A role for complexes of 79 Simsek M, Al-Bulushi T, Shanmugakonar M, Al-Barwani HS,
survival of motor neurons (SMN) protein with gemins and profilin Bayoumi R. Allele-specific amplification of exon 7 in the survival
in neurite-like cytoplasmic extensions of cultured nerve cells. motor neuron (SMN) genes for molecular diagnosis of spinal
Exp Cell Res 2005; 309: 185–97. muscular atrophy. Genet Test 2003; 7: 325–27.
58 Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G. 80 Wang CH, Carter TA, Das K, et al. Extensive DNA deletion
Axonal-SMN (a-SMN), a protein isoform of the survival motor associated with severe disease alleles on spinal muscular atrophy
neuron gene, is specifically involved in axonogenesis. homologues. Ann Neurol 1997; 42: 41–49.
Proc Natl Acad Sci USA 2007; 104: 1959–64. 81 Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B.
59 Rajendra TK, Gonsalvez GB, Walker MP, Shpargel KB, Salz HK, Quantitative analyses of SMN1 and SMN2 based on real-time
Matera AG. A Drosophila melanogaster model of spinal muscular lightCycler PCR: fast and highly reliable carrier testing and
atrophy reveals a function for SMN in striated muscle. J Cell Biol prediction of severity of spinal muscular atrophy. Am J Hum Genet
2007; 176: 831–41. 2002; 70: 358–68.
www.thelancet.com Vol 371 June 21, 2008 2131
Seminar
82 McAndrew PE, Parsons DW, Simard LR, et al. Identification of 104 Jarecki J, Chen X, Bernardino A, et al. Diverse small-molecule
proximal spinal muscular atrophy carriers and patients by analysis modulators of SMN expression found by high-throughput
of SMNT and SMNC gene copy number. Am J Hum Genet 1997; compound screening: early leads towards a therapeutic for spinal
60: 1411–22. muscular atrophy. Hum Mol Genet 2005; 14: 2003–18.
83 Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K. Genomic 105 Mattis VB, Rai R, Wang J, Chang CW, Coady T, Lorson CL. Novel
variation and gene conversion in spinal muscular atrophy: aminoglycosides increase SMN levels in spinal muscular atrophy
implications for disease process and clinical phenotype. fibroblasts. Hum Genet 2006; 120: 589–601.
Am J Hum Genet 1997; 61: 40–50. 106 Wolstencroft EC, Mattis V, Bajer AA, Young PJ, Lorson CL.
84 Carter TA, Bonnemann CG, Wang CH, et al. A multicopy A non-sequence-specific requirement for SMN protein activity: the
transcription-repair gene, BTF2p44, maps to the SMA region and role of aminoglycosides in inducing elevated SMN protein levels.
demonstrates SMA associated deletions. Hum Mol Genet 1997; Hum Mol Genet 2005; 14: 1199–210.
6: 229–36. 107 Lunn MR, Root DE, Martino AM, et al. Indoprofen upregulates the
85 Scharf JM, Endrizzi MG, Wetter A, et al. Identification of a survival motor neuron protein through a cyclooxygenase-
candidate modifying gene for spinal muscular atrophy by independent mechanism. Chem Biol 2004; 11: 1489–93.
comparative genomics. Nat Genet 1998; 20: 83–86. 108 Waza M, Adachi H, Katsuno M, Minamiyama M, Tanaka F,
86 Lefebvre S, Burlet P, Liu Q, et al. Correlation between severity and Sobue G. Alleviating neurodegeneration by an anticancer agent:
SMN protein level in spinal muscular atrophy. Nat Genet 1997; an Hsp90 inhibitor (17-AAG). Ann N Y Acad Sci 2006; 1086: 21–34.
16: 265–69. 109 Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B.
87 Scheffer H, Cobben JM, Mensink RG, Stulp RP, van der Steege G, Htra2-beta 1 stimulates an exonic splicing enhancer and can restore
Buys CH. SMA carrier testing: validation of hemizygous SMN full-length SMN expression to survival motor neuron 2 (SMN2).
exon 7 deletion test for the identification of proximal spinal Proc Natl Acad Sci USA 2000; 97: 9618–23.
muscular atrophy carriers and patients with a single allele 110 Pazin MJ, Kadonaga JT. What’s up and down with histone
deletion. Eur J Hum Genet 2000; 8: 79–86. deacetylation and transcription? Cell 1997; 89: 325–28.
88 Saugier-Veber P, Drouot N, Lefebvre S, et al. Detection of 111 Gilbert J, Baker SD, Bowling MK, et al. A phase I dose escalation
heterozygous SMN1 deletions in SMA families using a and bioavailability study of oral sodium phenylbutyrate in patients
simple fluorescent multiplex PCR method. J Med Genet 2001; with refractory solid tumor malignancies. Clin Cancer Res 2001;
38: 240–43. 7: 2292–300.
89 Chen KL, Wang YL, Rennert H, et al. Duplications and de novo 112 Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase
deletions of the SMNt gene demonstrated by fluorescence-based inhibitors. Adv Cancer Res 2004; 91: 137–68.
carrier testing for spinal muscular atrophy. Am J Med Genet 1999; 113 Hahnen E, Eyupoglu IY, Brichta L, et al. In vitro and ex vivo
85: 463–69. evaluation of second-generation histone deacetylase inhibitors for
90 Ogino S, Wilson RB. SMN dosage analysis and risk assessment for the treatment of spinal muscular atrophy. J Neurochem 2006;
spinal muscular atrophy. Am J Hum Genet 2002; 70: 1596–98. 98: 193–202.
91 Kesari A, Rennert H, Leonard DG, Phadke SR, Mittal B. Prenatal 114 Tsuji N, Kobayashi M, Nagashima K, Wakisaka Y, Koizumi K.
diagnosis of spinal muscular atrophy: Indian scenario. Prenat Diagn A new antifungal antibiotic, trichostatin. J Antibiot 1976; 29: 1–6.
2005; 25: 641–44. 115 Avila AM, Burnett BG, Taye AA, et al. Trichostatin A increases SMN
92 Wu T, Ding XS, Li WL, Yao J, Deng XX. Prenatal diagnosis of spinal expression and survival in a mouse model of spinal muscular
muscular atrophy in Chinese by genetic analysis of fetal cells. atrophy. J Clin Invest 2007; 117: 659–71.
Chinese Med J 2005; 118: 1274–77. 116 Stutzmann J, Wahl F, Pratt J, et al. Neuroprotective profile of
93 Beroud C, Karliova M, Bonnefont JP, et al. Prenatal diagnosis of riluzole in in vivo models of acute neurodegenerative disease.
spinal muscular atrophy by genetic analysis of circulating fetal cells. CNS Drug Rev 1997; 3: 83–101.
Lancet 2003; 361: 1013–14. 117 Haddad H, Cifuentes-Diaz C, Miroglio A, Roblot N, Joshi V,
94 Burlet P, Frydman N, Gigarel N, et al. Improved single-cell protocol Melki J. Riluzole attenuates spinal muscular atrophy disease
for preimplantation genetic diagnosis of spinal muscular atrophy. progression in a mouse model. Muscle Nerve 2003; 28: 432–37.
Fertil Steril 2005; 84: 734–39. 118 Angelozzi C, Borgo F, Tiziano FD, Martella A, Neri G, Brahe C.
95 Daniels G, Pettigrew R, Thornhill A, et al. Six unaffected livebirths Salbutamol increases SMN mRNA and protein levels in spinal
following preimplantation diagnosis for spinal muscular atrophy. muscular atrophy cells. J Med Genet 2008; 45: 29–31.
Mol Hum Reprod 2001; 7: 995–1000. 119 Yuo CY, Lin HH, Chang YS, Yang WK, Chang JG. 5-(N-ethyl-N-
96 Girardet A, Fernandez C, Claustres M. Efficient strategies for isopropyl)-amiloride enhances SMN2 exon 7 inclusion and protein
preimplantation genetic diagnosis of spinal muscular atrophy. expression in spinal muscular atrophy cells. Ann Neurol 2008;
Fertil Steril 2007; published online October 22. 63: 26–34.
DOI:10.1016/j.fertnstert.2007.07.1305. 120 Sakla MS, Lorson CL. Induction of full-length survival motor
97 Pyatt RE, Prior TW. A feasibility study for the newborn screening of neuron by polyphenol botanical compounds. Hum Genet 2008;
spinal muscular atrophy. Genet Med 2006; 8: 428–37. 122: 635–43.
98 Andreassi C, Jarecki J, Zhou J, et al. Aclarubicin treatment restores 121 Lorson CL, Strasswimmer J, Yao JM, et al. SMN oligomerization
SMN levels to cells derived from type I spinal muscular atrophy defect correlates with spinal muscular atrophy severity. Nat Genet
patients. Hum Mol Genet 2001; 10: 2841–49. 1998; 19: 63–66.
99 Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. 122 Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN
Treatment of spinal muscular atrophy by sodium butyrate. complex, an assemblyosome of ribonucleoproteins.
Proc Natl Acad Sci USA 2001; 98: 9808–13. Curr Opin Cell Biol 2002; 14: 305–12.
100 Brichta L, Hofmann Y, Hahnen E, et al. Valproic acid increases the 123 Hua Y, Zhou J. Modulation of SMN nuclear foci and
SMN2 protein level: a well-known drug as a potential therapy for cytoplasmic localization by its C-terminus. Cell Mol Life Sci 2004;
spinal muscular atrophy. Hum Mol Genet 2003; 12: 2481–89. 61: 2658–63.
101 Sumner CJ, Huynh TN, Markowitz JA, et al. Valproic acid increases 124 Howard MT, Anderson CB, Fass U, et al. Readthrough of
SMN levels in spinal muscular atrophy patient cells. Ann Neurol dystrophin stop codon mutations induced by aminoglycosides.
2003; 54: 647–54. Ann Neurol 2004; 55: 422–26.
102 Majumder S, Varadharaj S, Ghoshal K, Monani U, Burghes AH, 125 Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR.
Jacob ST. Identification of a novel cyclic AMP-response element Enhancement of SMN2 exon 7 inclusion by antisense
(CRE-II) and the role of CREB-1 in the cAMP-induced expression of the oligonucleotides targeting the exon. PLoS Biol 2007; 5: e73.
survival motor neuron (SMN) gene. J Biol Chem 2004; 279: 14803–11. 126 Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F.
103 Andreassi C, Angelozzi C, Tiziano FD, et al. Phenylbutyrate Bifunctional antisense oligonucleotides provide a trans-acting
increases SMN expression in vitro: relevance for treatment of spinal splicing enhancer that stimulates SMN2 gene expression in patient
muscular atrophy. Eur J Hum Genet 2004; 12: 59–65. fibroblasts. Proc Natl Acad Sci USA 2003; 100: 4114–19.
2132 www.thelancet.com Vol 371 June 21, 2008
Seminar
127 Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, 144 Feng W, Gubitz AK, Wan L, et al. Gemins modulate the expression
Dean NM. Modification of MyD88 mRNA splicing and inhibition of and activity of the SMN complex. Hum Mol Genet 2005;
IL-1beta signaling in cell culture and in mice with a 14: 1605–11.
2’-O-methoxyethyl-modified oligonucleotide. J Immunol 2006; 145 Azzouz M, Le T, Ralph GS, et al. Lentivector-mediated SMN
176: 3652–61. replacement in a mouse model of spinal muscular atrophy.
128 Cartegni L, Krainer AR. Correction of disease-associated exon J Clin Invest 2004; 114: 1726–31.
skipping by synthetic exon-speific activators. Nat Struct Biol 2003; 146 Coady TH, Shababi M, Tullis GE, Lorson CL. Restoration of SMN
10: 120–25. function: delivery of a trans-splicing RNA re-directs SMN2
129 DiMatteo D, Calahan S, Kmiec EB. Genetic conversion of an pre-mRNA splicing. Mol Ther 2007; 15: 1471–78.
SMN2 gene to SMN1: a novel approach to the treatment of spinal 147 Lesbordes JC, Cifuentes-Diaz C, Miroglio A, et al. Therapeutic
muscular atrophy. Exper Cell Res 2008; 314: 878–86. benefits of cardiotrophin-1 gene transfer in a mouse model of spinal
130 Bertini E, Burghes A, Bushby K, et al. 134th ENMC International muscular atrophy. Hum Mol Genet 2003; 12: 1233–39.
Workshop: outcome measures and treatment of spinal muscular 148 Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed
atrophy, 11–13 February 2005, Naarden, The Netherlands. differentiation of embryonic stem cells into motor neurons. Cell
Neuromuscul Disord 2005; 15: 802–16. 2002; 110: 385–97.
131 Kinali M, Mercuri E, Main M, et al. Pilot trial of albuterol in spinal 149 Wilson PG, Cherry JJ, Schwamberger S, et al. An SMA project
muscular atrophy. Neurol 2002; 59: 609–10. report: neural cell-based assays derived from human embryonic
132 Takeuchi Y, Miyanomae Y, Komatsu H, et al. Efficacy of stem cells. Stem Cells Dev 2007; 27: 1027–41.
thyrotropin-releasing hormone in the treatment of spinal muscular 150 Kerr DA, Llado J, Shamblott MJ, et al. Human embryonic germ cell
atrophy. J Child Neurol 1994; 9: 287–89. derivatives facilitate motor recovery of rats with diffuse motor
133 Brahe C, Vitali T, Tiziano FD, et al. Phenylbutyrate increases SMN neuron injury. J Neurosci 2003; 23: 5131–40.
gene expression in spinal muscular atrophy patients. 151 Harper JM, Krishnan C, Darman JS, et al. Axonal growth of
Eur J Hum Genet 2005; 13: 256–59. embryonic stem cell-derived motoneurons in vitro and in
134 Mercuri E, Bertini E, Messina S, et al. Pilot trial of phenylbutyrate motoneuron-injured adult rats. Proc Natl Acad Sci USA 2004;
in spinal muscular atrophy. Neuromuscul Disord 2004; 14: 130–35. 101: 7123–28.
135 Mercuri E, Bertini E, Messina S, et al. Randomized, double-blind, 152 Kaufmann P, Muntoni F. Issues in SMA clinical trial design: The
placebo-controlled trial of phenylbutyrate in spinal muscular International Coordinating Committee (ICC) for SMA
atrophy. Neurol 2007; 68: 51–55. Subcommittee on SMA Clinical Trial Design. Neuromuscul Disord
136 Miller RG, Moore DH, Dronsky V, et al. A placebo-controlled trial of 2007; 17: 499–505.
gabapentin in spinal muscular atrophy. J Neurol Sci 2001; 191: 127–31. 153 Merlini L, Mazzone ES, Solari A, Morandi L. Reliability of
137 Brichta L, Holker I, Haug K, Klockgether T, Wirth B. In vivo hand-held dynamometry in spinal muscular atrophy. Muscle Nerve
activation of SMN in spinal muscular atrophy carriers and patients 2002; 26: 64–70.
treated with valproate. Ann Neurol 2006; 59: 970–75. 154 Iannaccone ST. Outcome measures for pediatric spinal muscular
138 Weihl CC, Connolly AM, Pestronk A. Valproate may improve atrophy. Arch Neurol 2002; 59: 1445–50.
strength and function in patients with type III/IV spinal muscle 155 Iannaccone ST, Hynan LS. Reliability of 4 outcome measures in
atrophy. Neurol 2006; 67: 500–501. pediatric spinal muscular atrophy. Arch Neurol 2003; 60: 1130–36.
139 Russman BS, Iannaccone ST, Samaha FJ. A phase 1 trial of riluzole 156 Krosschell KJ, Maczulski JA, Crawford TO, Scott C, Swoboda KJ.
in spinal muscular atrophy. Arch Neurol 2003; 60: 1601–03. A modified Hammersmith functional motor scale for use in
140 Merlini L, Solari A, Vita G, et al. Role of gabapentin in spinal multi-center research on spinal muscular atrophy.
muscular atrophy: results of a multicenter, randomized Italian Neuromuscul Disord 2006; 16: 417–26.
study. J Child Neurol 2003; 18: 537–41. 157 Sumner CJ, Kolb SJ, Harmison GG, et al. SMN mRNA and protein
141 Liang WC, Yuo CY, Chang JG. The effect of hydroxyurea in spinal levels in peripheral blood: biomarkers for SMA clinical trials. Neurol
muscular atrophy cells and patients. J Neurol Sci 2008; 268: 87–94. 2006; 66: 1067–73.
142 Tzeng AC, Cheng J, Fryczynski H, et al. A study of 158 Simard LR, Belanger MC, Morissette S, Wride M, Prior TW,
thyrotropin-releasing hormone for the treatment of spinal muscular Swoboda KJ. Preclinical validation of a multiplex real-time assay to
atrophy: a preliminary report. Am J Phys Med Rehabil 2000; 79: 435–40. quantify SMN mRNA in patients with SMA. Neurol 2007;
143 Lunn MR, Stockwell BR. Chemical genetics and orphan genetic 68: 451–56.
diseases. Chem Biol 2005; 12: 1063–73.
www.thelancet.com Vol 371 June 21, 2008 2133
You might also like
- Neurology Multiple Choice Questions With Explanations: Volume IFrom EverandNeurology Multiple Choice Questions With Explanations: Volume IRating: 4 out of 5 stars4/5 (7)
- Alternative SplicingDocument356 pagesAlternative SplicingChalitha somarathne100% (3)
- Fast Facts: Diagnosing Amyotrophic Lateral Sclerosis: Clinical wisdom to facilitate faster diagnosisFrom EverandFast Facts: Diagnosing Amyotrophic Lateral Sclerosis: Clinical wisdom to facilitate faster diagnosisNo ratings yet
- ATROFIA MUSCULAR ESPINAL Lancet 2008Document14 pagesATROFIA MUSCULAR ESPINAL Lancet 2008Sebastián Silva SotoNo ratings yet
- Case Report: Coexistence of Ankylosing Spondylitis and Neurofibromatosis Type 1Document4 pagesCase Report: Coexistence of Ankylosing Spondylitis and Neurofibromatosis Type 1Lupita Mora LobacoNo ratings yet
- Benefits of Robotic Devices in Medical Rehabilitation of A Case With Neurofibromatosis Type1Document5 pagesBenefits of Robotic Devices in Medical Rehabilitation of A Case With Neurofibromatosis Type1Cioara IleanaNo ratings yet
- Congenital Muscular Dystrophies and Congenital.8 PDFDocument26 pagesCongenital Muscular Dystrophies and Congenital.8 PDFlithium2203No ratings yet
- Intensive Care Unit-Acquired WeaknessDocument13 pagesIntensive Care Unit-Acquired WeaknessAnonymous 3VkEwIsXRNo ratings yet
- Myoclonus With DementiaDocument8 pagesMyoclonus With DementiaSantosh DashNo ratings yet
- Amyotrophic Lateral SclerosisDocument11 pagesAmyotrophic Lateral SclerosisAnomalie12345No ratings yet
- Romero 2013Document16 pagesRomero 2013Rizki Nandasari SulbahriNo ratings yet
- A Review On Spinal Muscular Atrophy Clinical Classification, Etiology, Diagnosis and TreatmentDocument4 pagesA Review On Spinal Muscular Atrophy Clinical Classification, Etiology, Diagnosis and TreatmentEditor IJTSRDNo ratings yet
- Isolated Propriospinal Myoclonus As A Presentation of Cervical Myelopathy.Document2 pagesIsolated Propriospinal Myoclonus As A Presentation of Cervical Myelopathy.arturomarticarvajalNo ratings yet
- 2 - Debilidad AgudaDocument15 pages2 - Debilidad AgudaMarian ZeaNo ratings yet
- (2022) - Frozen Shoulder ReviewDocument16 pages(2022) - Frozen Shoulder ReviewCristobal LopezNo ratings yet
- Neuromuscular DisordersDocument12 pagesNeuromuscular DisordersZed HarrisNo ratings yet
- Transverse Myelitis: Clinical PracticeDocument9 pagesTransverse Myelitis: Clinical PracticearnabNo ratings yet
- Spinal Muscular Atrophy: Stephen J. Kolb,, John T. KisselDocument16 pagesSpinal Muscular Atrophy: Stephen J. Kolb,, John T. KisselZouhour MiladiNo ratings yet
- A Review of Lumbar Radiculopathy, Diagnosis, and TreatmentDocument7 pagesA Review of Lumbar Radiculopathy, Diagnosis, and TreatmentMatthew PhillipsNo ratings yet
- Evaluating Lumbar Spine 582-589Document8 pagesEvaluating Lumbar Spine 582-589Mansour ZamaniNo ratings yet
- Amyotrophic Lateral SclerosisDocument9 pagesAmyotrophic Lateral SclerosisCaroline ItnerNo ratings yet
- Jurnal SpondylosisDocument7 pagesJurnal SpondylosisM Hafiz MoulviNo ratings yet
- Neuromuscular Diseases PDFDocument205 pagesNeuromuscular Diseases PDFsalmazzNo ratings yet
- Osteochondroses: Franklin Danger, MD Christopher Wasyliw, MD Laura Varich, MDDocument7 pagesOsteochondroses: Franklin Danger, MD Christopher Wasyliw, MD Laura Varich, MDadrianNo ratings yet
- Hercules Baby - A Rare Case Presentation of Congenital MyopathyDocument4 pagesHercules Baby - A Rare Case Presentation of Congenital MyopathyInternational Journal of Innovative Science and Research TechnologyNo ratings yet
- En A04v86n4Document10 pagesEn A04v86n4Florentina NastaseNo ratings yet
- RRL - Scoliosis - Etiologic StudiesDocument6 pagesRRL - Scoliosis - Etiologic StudiesShao Anunciacion0% (1)
- Ogino and Wilson 2004Document16 pagesOgino and Wilson 2004akshayajainaNo ratings yet
- Case Study Myasthenia GravisDocument9 pagesCase Study Myasthenia GravisYow Mabalot100% (1)
- CV 20231223 32653A702 HonbunDocument7 pagesCV 20231223 32653A702 HonbunGrc hashtagNo ratings yet
- Preferred Examination: Spondylitis Ankylosing SpondylitisDocument6 pagesPreferred Examination: Spondylitis Ankylosing SpondylitischrisantyNo ratings yet
- A Pattern Recognition Approach To Myopathy.15Document24 pagesA Pattern Recognition Approach To Myopathy.15Maryam NabNo ratings yet
- Algoritmo Diferencial de La Ataxia Sensitiva en Mielopatia Por Cobre Secundaria A CeliaquiaDocument7 pagesAlgoritmo Diferencial de La Ataxia Sensitiva en Mielopatia Por Cobre Secundaria A CeliaquiaFarid Santiago Abedrabbo LombeydaNo ratings yet
- Algoritmo Diferencial de Debilidad Proximal en Paciente Con Miopatia Adquirida Por Nemalina.Document7 pagesAlgoritmo Diferencial de Debilidad Proximal en Paciente Con Miopatia Adquirida Por Nemalina.Farid Santiago Abedrabbo LombeydaNo ratings yet
- Clinical and Pathologic Aspects of Congenital Myopathies: Ikuya Nonaka MDDocument8 pagesClinical and Pathologic Aspects of Congenital Myopathies: Ikuya Nonaka MDEvlin KoharNo ratings yet
- Cervical SpondylosisDocument10 pagesCervical Spondylosistaufiq duppa duppaaNo ratings yet
- Marinesco-Sjo Gren Syndrome - 2013Document5 pagesMarinesco-Sjo Gren Syndrome - 2013Irfan RazaNo ratings yet
- Tob 12.2Document28 pagesTob 12.2LT DRAGONXNo ratings yet
- Ankylosing Spondylarthritis PDFDocument3 pagesAnkylosing Spondylarthritis PDFMa OlayaNo ratings yet
- Congenital Myopathies: Clinical Phenotypes and New Diagnostic ToolsDocument16 pagesCongenital Myopathies: Clinical Phenotypes and New Diagnostic ToolsRizki Nandasari SulbahriNo ratings yet
- Miopati - Referensi MiopatiDocument25 pagesMiopati - Referensi MiopatiKelvin Theandro GotamaNo ratings yet
- Fas CaseDocument2 pagesFas Caseanon_378668477No ratings yet
- Arthrogryposis Causes, Consequences and Clinical Course in Amyoplasia and Distal ArthrogryposisDocument64 pagesArthrogryposis Causes, Consequences and Clinical Course in Amyoplasia and Distal ArthrogryposisJanina DrăgoiNo ratings yet
- Cervical Spondylosis and Neck PainDocument5 pagesCervical Spondylosis and Neck PainIsaac AlemanNo ratings yet
- Care PlanDocument20 pagesCare PlanamantelNo ratings yet
- Atrfoia Espinal MioclonicDocument18 pagesAtrfoia Espinal MioclonicmmblozsnoNo ratings yet
- Protrusio Acetabuli Diagnosis and TreatmentDocument10 pagesProtrusio Acetabuli Diagnosis and TreatmentFerney Leon BalceroNo ratings yet
- Spinal Muscular AtrophyDocument11 pagesSpinal Muscular AtrophyRahma NovitasariNo ratings yet
- Case Report of Intrafamilial Variability in Autosomal Recessive Centronuclear Myopathy Associated To A Novel BIN1 Stop MutationDocument6 pagesCase Report of Intrafamilial Variability in Autosomal Recessive Centronuclear Myopathy Associated To A Novel BIN1 Stop Mutationyael1991No ratings yet
- Maedica Art 18Document6 pagesMaedica Art 18Ersya MuslihNo ratings yet
- 03 Sarah OConnorDocument3 pages03 Sarah OConnorMateen ShukriNo ratings yet
- A Review of Lumbar Radiculopathy, Diagnosis, and TreatmentDocument7 pagesA Review of Lumbar Radiculopathy, Diagnosis, and TreatmentZuraidaNo ratings yet
- Distrofias Musculares 2019 MercuriDocument14 pagesDistrofias Musculares 2019 Mercurimaria palaciosNo ratings yet
- Idiopathic ScoliosisDocument12 pagesIdiopathic ScoliosisAlessandro BergantinNo ratings yet
- Spinal Muscular AthropyDocument13 pagesSpinal Muscular AthropyooccacNo ratings yet
- DownloadDocument15 pagesDownloadM Aqil GibranNo ratings yet
- (Nigel G. Laing) The Sarcomere and Skeletal Muscle PDFDocument246 pages(Nigel G. Laing) The Sarcomere and Skeletal Muscle PDFVladimir Pablo Morales QuevedoNo ratings yet
- Autoimmune and Paraneoplastic MyeloDocument12 pagesAutoimmune and Paraneoplastic MyeloSimply SamNo ratings yet
- Debilidad Neuromuscular Adquirida y Movilizacion Temprana en UTIDocument14 pagesDebilidad Neuromuscular Adquirida y Movilizacion Temprana en UTILucho FelicevichNo ratings yet
- HTTPS:WWW Ncbi NLM Nih gov:pmc:articles:PMC9144471:pdf:jpm-12-00808Document12 pagesHTTPS:WWW Ncbi NLM Nih gov:pmc:articles:PMC9144471:pdf:jpm-12-00808Ihsanul Ma'arifNo ratings yet
- Journal of Neurology and NeurocienceDocument49 pagesJournal of Neurology and NeurocienceInternational Medical PublisherNo ratings yet
- Genic-Balance Theory of Sex DeterminationDocument19 pagesGenic-Balance Theory of Sex Determinationstevensb05575% (4)
- Bioinformatic PracticeDocument4 pagesBioinformatic Practiceyungiang157No ratings yet
- 1 s2.0 S1044579X20302790 Main PDFDocument10 pages1 s2.0 S1044579X20302790 Main PDFgarrobosNo ratings yet
- Gene ConceptDocument80 pagesGene ConceptPrince HamdaniNo ratings yet
- How The Genes Are Regulated & Expressed?: AssignmentDocument17 pagesHow The Genes Are Regulated & Expressed?: AssignmentSuraiya Siraj NehaNo ratings yet
- Molecular Biology of The Cell, Sixth Edition Chapter 7: Control of Gene ExpressionDocument36 pagesMolecular Biology of The Cell, Sixth Edition Chapter 7: Control of Gene ExpressionIsmael Torres-Pizarro100% (2)
- 002 - 2018 - Splicing Mutations in Human Genetic Disorders - Examples Detection and ConfirmationDocument16 pages002 - 2018 - Splicing Mutations in Human Genetic Disorders - Examples Detection and ConfirmationAndrea Bermúdez QuinteroNo ratings yet
- In Silico Analysis of BRCA1 andDocument23 pagesIn Silico Analysis of BRCA1 and02213No ratings yet
- Unit-3 BioinformaticsDocument15 pagesUnit-3 Bioinformaticsp vmuraliNo ratings yet
- RNA Processing in Eukaryotes: January 15, 2021 Gaurab KarkiDocument5 pagesRNA Processing in Eukaryotes: January 15, 2021 Gaurab KarkiShahriar ShamimNo ratings yet
- Review Article: Hereditary Breast Cancer: The Era of New Susceptibility GenesDocument12 pagesReview Article: Hereditary Breast Cancer: The Era of New Susceptibility GenesSara MagoNo ratings yet
- Transcription Sample Old Essay PDFDocument7 pagesTranscription Sample Old Essay PDFOccamsRazorNo ratings yet
- Mutaciones BGM PDFDocument6 pagesMutaciones BGM PDFwaldo tapiaNo ratings yet
- Primer Design Using IDT Quest: Identifying NCBI Transcript & Associated PrimersDocument3 pagesPrimer Design Using IDT Quest: Identifying NCBI Transcript & Associated PrimersAqsa MuzammilNo ratings yet
- Han 2010Document14 pagesHan 2010julia rossoNo ratings yet
- Lesson Plan Day 1Document3 pagesLesson Plan Day 1Stephanie Ngooi Ming MingNo ratings yet
- Scientific Report 2007: 3D Glasses InsideDocument38 pagesScientific Report 2007: 3D Glasses Insidea4agarwal100% (5)
- Unit 2 Cells, Development, Biodiversity and ConservationDocument47 pagesUnit 2 Cells, Development, Biodiversity and ConservationEllane leeNo ratings yet
- 357 - Cell-Biology Physiology) DNA Transcription001Document12 pages357 - Cell-Biology Physiology) DNA Transcription001SodysserNo ratings yet
- News and Views: Advancing RNA-Seq AnalysisDocument3 pagesNews and Views: Advancing RNA-Seq AnalysisWilson Rodrigo Cruz FlorNo ratings yet
- Vanilloid (Capsaicin) Receptors in Health and Disease: Arpad Szallasi, MD, PHDDocument12 pagesVanilloid (Capsaicin) Receptors in Health and Disease: Arpad Szallasi, MD, PHDprift2No ratings yet
- Data RetrievalDocument17 pagesData RetrievalAyesha Khan50% (2)
- AV UWorld EOs (Rough Draft) - Data - Subject LandscapeDocument139 pagesAV UWorld EOs (Rough Draft) - Data - Subject Landscapesaeedfadaly1No ratings yet
- Gene Expression in EukaryotesDocument26 pagesGene Expression in EukaryotesArap DomNo ratings yet
- Myth of Junk DNA Notes (50p)Document50 pagesMyth of Junk DNA Notes (50p)siriusaz9398100% (1)
- Plant Grafting: PrimerDocument6 pagesPlant Grafting: Primerikhlas65No ratings yet
- Intelligence N GenesDocument208 pagesIntelligence N Genestoussman80No ratings yet
- Comparative Proteomics Kit I: Protein Profiler Module: Biotechnology ExplorerDocument50 pagesComparative Proteomics Kit I: Protein Profiler Module: Biotechnology ExplorerHidayat FadelanNo ratings yet
- RNA SplicingDocument8 pagesRNA SplicingDivya NarayanNo ratings yet