Professional Documents
Culture Documents
Data Import Structure Elucidation Interactive Data Browser Feature Table
Data Import Structure Elucidation Interactive Data Browser Feature Table
Uploaded by
Ignacio Pérez-VictoriaCopyright:
Available Formats
You might also like
- Manual - Cube Sugar - V02 - ENCNFRDEJP - 190104 PDFDocument36 pagesManual - Cube Sugar - V02 - ENCNFRDEJP - 190104 PDFIgnacio Pérez-Victoria100% (1)
- Asta Analytical MethodsDocument4 pagesAsta Analytical Methodsravivarmahyd8173No ratings yet
- Rivaroxaban Tablets Pharmeuropa 31.02EDocument4 pagesRivaroxaban Tablets Pharmeuropa 31.02EMariaNo ratings yet
- Unit VI Molecular Spectroscopy: Physical ChemistryDocument17 pagesUnit VI Molecular Spectroscopy: Physical ChemistryDewal Deshmukh100% (1)
- Hall of Fame: David Russell Recordings and EditionsDocument5 pagesHall of Fame: David Russell Recordings and EditionsIgnacio Pérez-Victoria100% (1)
- Modelbase System: A Distributed Model Database On The InternetDocument5 pagesModelbase System: A Distributed Model Database On The InternetKirupa KaranNo ratings yet
- Image Retrieval by ExamplesDocument15 pagesImage Retrieval by ExamplesHkh HkhNo ratings yet
- 0 MetaboAnalyst 5.0 OverviewDocument7 pages0 MetaboAnalyst 5.0 Overview유승하No ratings yet
- Performance and Analyses Using Two ETL Extraction Software SolutionsDocument4 pagesPerformance and Analyses Using Two ETL Extraction Software SolutionsVanya SinghalNo ratings yet
- SumitKumarDam - 09-05-2024Document21 pagesSumitKumarDam - 09-05-2024SUMITKUMARDAM[학생](대학원 인공지능학과) No ratings yet
- Entity Relationship Model: Pamantasan NG CabuyaoDocument10 pagesEntity Relationship Model: Pamantasan NG CabuyaoBien MedinaNo ratings yet
- Journal PublishersDocument4 pagesJournal Publishersgraphic designerNo ratings yet
- MetaboAnalyst 5.0 - Narrowing The Gap Between Raw Spectra and Func - 2Document11 pagesMetaboAnalyst 5.0 - Narrowing The Gap Between Raw Spectra and Func - 2Iqbal PrawiraNo ratings yet
- Ali ResumeDocument2 pagesAli Resumeaftab28No ratings yet
- © 2008 University of Delaware, All Rights Reserved Find Us On The Web atDocument2 pages© 2008 University of Delaware, All Rights Reserved Find Us On The Web atkannanmech87No ratings yet
- Poster SurfDocument1 pagePoster SurfEmilio FerraraNo ratings yet
- Week 1Document4 pagesWeek 1katrusampathvishal879No ratings yet
- Leveraging Lightweight Semantics For Search ImprovDocument3 pagesLeveraging Lightweight Semantics For Search ImprovJoffre ReynoldsNo ratings yet
- Weka Software ManualaDocument20 pagesWeka Software ManualaAswin Kumar ThanikachalamNo ratings yet
- DBMS PROJECT Overview - Roll No. 07 & 16Document9 pagesDBMS PROJECT Overview - Roll No. 07 & 16parth vermaNo ratings yet
- Automotive Embedded System SyllabusDocument5 pagesAutomotive Embedded System SyllabusgudduNo ratings yet
- Entity Extraction SystemDocument6 pagesEntity Extraction SystemUday SolNo ratings yet
- A Survey On Software Suites For Data Mining, Analytics and Knowledge DiscoveryDocument6 pagesA Survey On Software Suites For Data Mining, Analytics and Knowledge DiscoveryInternational Journal of computational Engineering research (IJCER)No ratings yet
- Symmetry: Webshell Attack Detection Based On A Deep Super LearnerDocument16 pagesSymmetry: Webshell Attack Detection Based On A Deep Super LearnerDevil izalNo ratings yet
- RE ProposalDocument25 pagesRE ProposalJiun sheng WONGNo ratings yet
- Semantic Product SearchDocument4 pagesSemantic Product SearchYaron RianyNo ratings yet
- CrossMedia: E-Science Collaboration Platform For Effective Multimedia ResearchDocument5 pagesCrossMedia: E-Science Collaboration Platform For Effective Multimedia ResearchAndras MicsikNo ratings yet
- A Modular Reconfigurable and Portable Framework For On-Board Data Processing Architecture and ApplicationsDocument6 pagesA Modular Reconfigurable and Portable Framework For On-Board Data Processing Architecture and ApplicationsBoul chandra GaraiNo ratings yet
- PolyAnalyst Overview ENGDocument31 pagesPolyAnalyst Overview ENGEugene SolomatinNo ratings yet
- Mol ViewerDocument7 pagesMol Viewerwonderopokuasante9No ratings yet
- Eisen PytorchDocument8 pagesEisen PytorchLuboš ŘehounekNo ratings yet
- 1 s2.0 S0169260721004880 MainDocument13 pages1 s2.0 S0169260721004880 MainmaketoNo ratings yet
- Information ExtractionDocument7 pagesInformation ExtractionBini Teflon AnkhNo ratings yet
- Sqmtools: Automated Processing and Visual Analysis of 'Omics Data With R and Anvi'ODocument11 pagesSqmtools: Automated Processing and Visual Analysis of 'Omics Data With R and Anvi'OshanmugapriyaNo ratings yet
- INCOSE IW09-MBSE - Initiative - SysMO - (d01FEB09-aRH-v1.4-sFINAL)Document42 pagesINCOSE IW09-MBSE - Initiative - SysMO - (d01FEB09-aRH-v1.4-sFINAL)Ralph HodgsonNo ratings yet
- Elizabeth Walkup, MacMalwareDocument5 pagesElizabeth Walkup, MacMalwareselavilizationNo ratings yet
- DB MinerDocument6 pagesDB MinerNishant KumarNo ratings yet
- Computers OverviewDocument3 pagesComputers Overviewfiw ahimNo ratings yet
- Patterns For Model-Driven Software-DevelopmentDocument50 pagesPatterns For Model-Driven Software-DevelopmentOtto F OttONo ratings yet
- Professional Program in Data Science and Machine Learning: - ML EngineerDocument4 pagesProfessional Program in Data Science and Machine Learning: - ML EngineerMANPREET SODHINo ratings yet
- Information ExtractionDocument8 pagesInformation ExtractionBini Teflon AnkhNo ratings yet
- Information ExtractionDocument8 pagesInformation ExtractionBini Teflon AnkhNo ratings yet
- Metaseek: A Content-Based Meta-Search Engine For ImagesDocument11 pagesMetaseek: A Content-Based Meta-Search Engine For ImagesEman AlkurdiNo ratings yet
- Dynamic Filtering and Prioritization of Static Code Analysis AlertsDocument2 pagesDynamic Filtering and Prioritization of Static Code Analysis Alertscagla.cengizNo ratings yet
- SIRIUS 4 - A Rapid Tool For Turning Tandem Mass Spectra Into Metabolite Structure InformationDocument10 pagesSIRIUS 4 - A Rapid Tool For Turning Tandem Mass Spectra Into Metabolite Structure InformationJovanderson JacksonNo ratings yet
- Imperial College London (MMG, Yg, Ac901) @doc - Ic.Ac - Uk Inforsense LTDDocument4 pagesImperial College London (MMG, Yg, Ac901) @doc - Ic.Ac - Uk Inforsense LTDNhek VibolNo ratings yet
- General Katalog Ditek JayaDocument52 pagesGeneral Katalog Ditek JayamedisoniantoNo ratings yet
- Information Systems Review of Literatures: II. System ArchitectureDocument3 pagesInformation Systems Review of Literatures: II. System ArchitectureLucman AbdulrachmanNo ratings yet
- Plantweb Optics Data Lake ComponentsDocument2 pagesPlantweb Optics Data Lake ComponentskEWQ 865kNo ratings yet
- Database Management System SKILLING PROJECT-Mojes Film Makers Plan of Action SNO Component Accomplishment Status RemarkDocument1 pageDatabase Management System SKILLING PROJECT-Mojes Film Makers Plan of Action SNO Component Accomplishment Status Remarkprudhvi chNo ratings yet
- Cdisc 2011 Dandamudi PDFDocument1 pageCdisc 2011 Dandamudi PDFKrishna DandamudiNo ratings yet
- Kim 2018Document18 pagesKim 2018Julian Rolando Avendaño PeñaNo ratings yet
- Knowledge Map Design of Bioethics Supporting 3D VisualizationDocument5 pagesKnowledge Map Design of Bioethics Supporting 3D VisualizationStefanyNo ratings yet
- Advance Excel COURSEDocument6 pagesAdvance Excel COURSEjahanviNo ratings yet
- Computer Physics Communications: Vei Wang, Nan Xu, Jin-Cheng Liu, Gang Tang, Wen-Tong GengDocument19 pagesComputer Physics Communications: Vei Wang, Nan Xu, Jin-Cheng Liu, Gang Tang, Wen-Tong Genglixiang RaoNo ratings yet
- Assignment No. 7: Shri Guru Gobind Singhji Institute of Engineering and Technology (SGGSIET), NandedDocument3 pagesAssignment No. 7: Shri Guru Gobind Singhji Institute of Engineering and Technology (SGGSIET), NandedAjay JadhavNo ratings yet
- (IJCST-V12I1P6) :kaushik Kashyap, Rinku Moni Borah, Priyanku Rahang, DR Bornali Gogoi, Prof. Nelson R VarteDocument5 pages(IJCST-V12I1P6) :kaushik Kashyap, Rinku Moni Borah, Priyanku Rahang, DR Bornali Gogoi, Prof. Nelson R VarteEighthSenseGroupNo ratings yet
- Data Warehouse Operational Architecture: Keywords: DB2, Oracle, Data Warehouse Data Warehouse EnvironmentDocument14 pagesData Warehouse Operational Architecture: Keywords: DB2, Oracle, Data Warehouse Data Warehouse EnvironmentRoberto GavidiaNo ratings yet
- Inputs To System DesignDocument8 pagesInputs To System Design555bsyadav555No ratings yet
- Efficient Structure-Informed Featurization and Property Prediction of Ordered, Dilute, and Random Atomic StructuresDocument23 pagesEfficient Structure-Informed Featurization and Property Prediction of Ordered, Dilute, and Random Atomic Structuresdemoc29381hkgsc12343No ratings yet
- ML Training PDFDocument6 pagesML Training PDFshrestha3902No ratings yet
- Conclusion: So Store Manager: It Stores The Sos, Sos MetadataDocument1 pageConclusion: So Store Manager: It Stores The Sos, Sos MetadataSrinivasNo ratings yet
- NASA Constellation Program Ontologies Ralph Hodgson 20080320Document48 pagesNASA Constellation Program Ontologies Ralph Hodgson 20080320Ralph HodgsonNo ratings yet
- UCS RM Intro-EtlDocument30 pagesUCS RM Intro-Etlnur ashfaralianaNo ratings yet
- DATA MINING and MACHINE LEARNING. PREDICTIVE TECHNIQUES: ENSEMBLE METHODS, BOOSTING, BAGGING, RANDOM FOREST, DECISION TREES and REGRESSION TREES.: Examples with MATLABFrom EverandDATA MINING and MACHINE LEARNING. PREDICTIVE TECHNIQUES: ENSEMBLE METHODS, BOOSTING, BAGGING, RANDOM FOREST, DECISION TREES and REGRESSION TREES.: Examples with MATLABNo ratings yet
- Cord-Based Microfluidic Chips As A Platform For ELISA and Glucose AssaysDocument11 pagesCord-Based Microfluidic Chips As A Platform For ELISA and Glucose AssaysIgnacio Pérez-VictoriaNo ratings yet
- Specs PalosDocument3 pagesSpecs PalosIgnacio Pérez-VictoriaNo ratings yet
- Romance D'amour: Arranged Dang Thao NguyenDocument6 pagesRomance D'amour: Arranged Dang Thao NguyenIgnacio Pérez-VictoriaNo ratings yet
- Derive Lab Manual-1Document248 pagesDerive Lab Manual-1Ignacio Pérez-VictoriaNo ratings yet
- Marine Drugs: New 1,4-Dienonesteroids From The Octocoral Dendronephthya SPDocument8 pagesMarine Drugs: New 1,4-Dienonesteroids From The Octocoral Dendronephthya SPIgnacio Pérez-VictoriaNo ratings yet
- Smash 2019 ProgramDocument225 pagesSmash 2019 ProgramIgnacio Pérez-VictoriaNo ratings yet
- Marinedrugs 17 00457Document17 pagesMarinedrugs 17 00457Ignacio Pérez-VictoriaNo ratings yet
- Fender Blacktop & Modern Player ElectricsDocument5 pagesFender Blacktop & Modern Player ElectricsIgnacio Pérez-VictoriaNo ratings yet
- CHAPTER 1-Reading Music and Tablature (TAB) : Intermediate Acoustic GuitarDocument6 pagesCHAPTER 1-Reading Music and Tablature (TAB) : Intermediate Acoustic GuitarIgnacio Pérez-VictoriaNo ratings yet
- Metabolites: The Metarbolomics Toolbox in Bioconductor and BeyondDocument55 pagesMetabolites: The Metarbolomics Toolbox in Bioconductor and BeyondIgnacio Pérez-VictoriaNo ratings yet
- Marine Drugs: Bioactive Bromotyrosine-Derived Alkaloids From The Polynesian Sponge Suberea IanthelliformisDocument16 pagesMarine Drugs: Bioactive Bromotyrosine-Derived Alkaloids From The Polynesian Sponge Suberea IanthelliformisIgnacio Pérez-VictoriaNo ratings yet
- Johann Sebastian BachDocument6 pagesJohann Sebastian BachIgnacio Pérez-VictoriaNo ratings yet
- Metabolites 09 00165 v2Document15 pagesMetabolites 09 00165 v2Ignacio Pérez-VictoriaNo ratings yet
- Marine Drugs: Synthesis and Antitumor Activity Evaluation of Compounds Based On ToluquinolDocument16 pagesMarine Drugs: Synthesis and Antitumor Activity Evaluation of Compounds Based On ToluquinolIgnacio Pérez-VictoriaNo ratings yet
- Marinedrugs 17 00402Document16 pagesMarinedrugs 17 00402Ignacio Pérez-VictoriaNo ratings yet
- Molecules: A Self-Healing and Shape Memory Polymer That Functions at Body TemperatureDocument12 pagesMolecules: A Self-Healing and Shape Memory Polymer That Functions at Body TemperatureIgnacio Pérez-VictoriaNo ratings yet
- Article MoleculesDocument11 pagesArticle MoleculesIgnacio Pérez-VictoriaNo ratings yet
- Marinedrugs 17 00167Document10 pagesMarinedrugs 17 00167Ignacio Pérez-VictoriaNo ratings yet
- Molecules 24 03221Document12 pagesMolecules 24 03221Ignacio Pérez-VictoriaNo ratings yet
- Article MetabolitesDocument25 pagesArticle MetabolitesIgnacio Pérez-VictoriaNo ratings yet
- Beethoven: Selected Works Transcribed For GuitarDocument6 pagesBeethoven: Selected Works Transcribed For GuitarIgnacio Pérez-VictoriaNo ratings yet
- A Systematic Review of Recently Reported Marine Derived Natural Product Kinase InhibitorsDocument38 pagesA Systematic Review of Recently Reported Marine Derived Natural Product Kinase InhibitorsIgnacio Pérez-VictoriaNo ratings yet
- James Ashary Habibie - 191501071 - Jurnal Kafein (ACC NILAI)Document72 pagesJames Ashary Habibie - 191501071 - Jurnal Kafein (ACC NILAI)Habibie TanjungNo ratings yet
- Diffraction GratingDocument9 pagesDiffraction GratingRajaswi BeleNo ratings yet
- LC FinalDocument11 pagesLC FinalCharlez UmerezNo ratings yet
- 1.HPLC Presentation MITDocument58 pages1.HPLC Presentation MITDuc Nhon LENo ratings yet
- Thin Layer ChromatographyDocument8 pagesThin Layer ChromatographyIsabel RinconNo ratings yet
- UV Vis Spectrophotometry Selection GuideDocument9 pagesUV Vis Spectrophotometry Selection GuideDolphingNo ratings yet
- Chapter 1 - Introduction To Spectrometric MethodsDocument57 pagesChapter 1 - Introduction To Spectrometric MethodsFarvin FleetNo ratings yet
- Atomic Absorption SpectrometryDocument64 pagesAtomic Absorption Spectrometryanilrockzzz786No ratings yet
- Forensic Sci Bibliography, ExplosivesDocument4 pagesForensic Sci Bibliography, ExplosivesSusan ColemanNo ratings yet
- Protocolo Purificacion Desde GelDocument3 pagesProtocolo Purificacion Desde GelAriel ArayaNo ratings yet
- Bio 120 Lab Ex 4 PostlabDocument2 pagesBio 120 Lab Ex 4 PostlabSunflowerNo ratings yet
- PRECIPITATIONDocument24 pagesPRECIPITATIONkekoacybelleNo ratings yet
- FTIR ReportDocument15 pagesFTIR Reportapi-3813659100% (10)
- PreviewpdfDocument68 pagesPreviewpdfJoel Ccallo HuaquistoNo ratings yet
- AguerDocument23 pagesAguerHuruk BrosnanNo ratings yet
- Lab.7 عقاقير ثانيDocument9 pagesLab.7 عقاقير ثانيهاني عقيل حسين جوادNo ratings yet
- Spectrochemical Analysis by Atomic Absorption and Emission - Lajunen 2nd Edition 2004Document360 pagesSpectrochemical Analysis by Atomic Absorption and Emission - Lajunen 2nd Edition 2004Goretti Arvizu100% (1)
- Assignment On HPLC and TLC - PDF - Pharmacognosy & PhytochemistryDocument9 pagesAssignment On HPLC and TLC - PDF - Pharmacognosy & PhytochemistryMr HotmasterNo ratings yet
- Lista Peças 2Document5 pagesLista Peças 2mardonio andradeNo ratings yet
- Gatorade Beer's Law Lab-Chem 4-1Document5 pagesGatorade Beer's Law Lab-Chem 4-1Mark Cliffton BadlonNo ratings yet
- Simplified Enrichment Method Using NeedlEx. Application Note (Shimadzu)Document2 pagesSimplified Enrichment Method Using NeedlEx. Application Note (Shimadzu)Maikel Perez NavarroNo ratings yet
- Analytical Techniques in Pharmaceutical Analysis ADocument14 pagesAnalytical Techniques in Pharmaceutical Analysis AKeshavVashistha100% (1)
- Gel Filtration Selection GuideDocument1 pageGel Filtration Selection GuideDolphingNo ratings yet
- Affinity of LDH LabDocument5 pagesAffinity of LDH LabZoe A GarwoodNo ratings yet
- Combined Spectra ProblemsDocument11 pagesCombined Spectra ProblemsUsama khanNo ratings yet
- ChromatographyDocument28 pagesChromatographyyashaviNo ratings yet
- Protein Analysis WorkflowDocument37 pagesProtein Analysis WorkflowKurdianto MSiNo ratings yet
Data Import Structure Elucidation Interactive Data Browser Feature Table
Data Import Structure Elucidation Interactive Data Browser Feature Table
Uploaded by
Ignacio Pérez-VictoriaOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Data Import Structure Elucidation Interactive Data Browser Feature Table
Data Import Structure Elucidation Interactive Data Browser Feature Table
Uploaded by
Ignacio Pérez-VictoriaCopyright:
Available Formats
Small Molecule Discovery from LC-MS Data with Metaboseek
Maximilian J. Helf*, Bennett W. Fox, Frank C. Schroeder
Boyce Thompson Institute and Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York, USA.
*Present address: Natural Products and Biomolecular Chemistry Unit, Novartis Institutes of BioMedical Research, Basel, Switzerland.
Contact: maximilian.helf@gmail.com
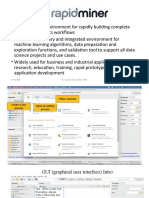
Data Import Feature Table Interactive Data Browser Structure Elucidation Peptide App Built From
Load feature tables
Navigation of data is centered on the feature table, a list
of molecular features, defined by at least m/z and retention
Browse, filter and annotate LC/MS features of interest.
Use powerful visualization tools to view and export
Use integrated tools to aid structure elucidation of molecular fea-
tures of interest. Save time by viewing the results in Metabo-
Metaboseek Modules
from other MS data External tools time (rt). Molecular features can be identified in MS data high-quality data plots. seek with quick access to the original raw data.
analysis tools or run
Import/ with Metaboseek’s xcms module or using external tools.
Customizable EIC Display Submit & View SIRIUS Jobs Metaboseek is built from individual modules that can be rear-
xcms analyses on xcms (Online) ranged easily into new shiny apps.
your MS data directly Plot extracted ion chromatograms (EICs) of molecular fea- Run SIRIUS³ analysis
from the Metaboseek MetaboAnalyst Export m/z rt {intensities, FC, p-value, ...} tures across files and arrange data in groups for rapid from the Metaboseek in-
interface.
mzMINE manual inspection terface and view results.
b2 b3 Use the PTModseek app Build a graphical user
LC/ HRMS 556.234 3.45 0 412 0 ... Export reports for individ- to annotate MS2 spectra interface for your data
Group 1 Group 2 ual molecular features, LL I
3.0e+07
of modified peptides analysis scripts
678.341 4.46 124 31 142 ... including their EIC and
SIRIUS fragmentation
XCMS 199.143 7.25 70 33 725 ... tree. Alternatively, open
227.1754+ ( 0.1)
340.2595+ ( 0.0)
500.2902+(−0.2)
658.3604+(−1.8)
873.4905+(−4.9)
990.5084+ ( 2.7)[1Me|1CAM]
1207.563+ ( 0.3)[1Me|3CAM]
1207.563+ ( 0.3)[1Me|2CAM]
1437.649+ ( 2.8)[1Me|2CAM]
1684.712+ ( 2.7)[1Me|3CAM]
1685.722+(−4.1)
786.453+ ( 1.5)
the analysis results in
Module
2.0e+07
MS Data Files Comparative Analysis SIRIUS GUI.
R
bn bn+1
O
Normalize feature intensities and run statisti- Label Finder N
H
N
N
cal analyses to compare sample groups for H H
[1CAM]
[1CAM]
[1CAM]
[1CAM]
Use the label finder module to detect stable
Parent O R
1.0e+07
feature-specific fold-change, t-test and y m+1 ym
isotope labeled compounds.
ANOVA. Import and export to MetaboAnalyst m/z b2 b3 b4 b6 b7 b8 b9 b10 b11 b13 b15 b16
b- and y- ions
Intensity
Relative Intensity
L L I C S A K S C C G E T S CWA C G
Accelerating Compound for advanced statistics. y5 y4 y3 y2
653.2164+ ( 0.9) [2CAM]
493.187+(−1.4) [1CAM]
307.1071+(−0.2) [1CAM]
236.0698+ ( 0.5) [1CAM]
Discovery C1
0
13
MS Data Reanalysis
11.8 12.0 12.2 12.4 11.8 12.0 12.2 12.4
13
C4
RT (min) RT (min)
13
C2 C3
13
C5
Comparative analysis of high resolution mass spectrometry (HRMS)
Reanalyze raw data for molecular features to
add information such as peak quality, exis-
MS/MS Browser 13
3.482e+04 RT: 4.496 min
y+2[1CAM]
100
View and compare mass spectra, including MS2 spectra Parent m/z: 1088.95398
data is a powerful approach to identify products and intermediates of
b+2
tence of custom fragmentation patterns MS2 m/z
belonging to individual nodes of a molecular network 80
biosynthetic pathways. However, making data analysis tools available data, or molecular formula database matches.
OH
Relative Intensity (%)
and moving between them is presenting a productivity bottleneck, 100
OH
572.2867 O 60
potentially leading to missed discovery opportunities. Custom Filters O
b+ +15[1Me|3CAM]
O
y+3[1CAM]
b+13[1Me|2CAM]
O O O
333.0607
b+11[1Me|2CAM]
HO
b+9[1Me|1CAM]
+ y4[1CAM]
P
40
N
b+8[1CAM]
Use results from comparative analysis, fea-
y++5[2CAM]
b6[1CAM]
b4[1CAM]
152.9965
To facilitate the identification of biologically relevant small molecules, HO
b+7[1CAM]
ture detection and MS data reanalysis to filter OH H
+
OH 20
336.1960
b+3
we recently introduced Metaboseek¹, an R/shiny-based, discovery-ori-
b16
for the features you want to investigate and
ented platform to extract differential features from HPLC-HRMS data
in a friendly graphical user interface. It allows you to rapidly inspect
add custom comments to molecular features. Feature-based Molecular Networking 0
500 1000 1500
410.2331 m/z
feature-specific data, find groups of structurally related com- Generate and browse molecular
0
networks based on MS2 similarity. Data shown here was previously published⁴ and reanalyzed with
pounds and assist compound identification and dereplication. Fold-change Peak Quality 100
Networks can also be exported to the PTModseek app, (included in the Metaboseek installer).
586.3027
Cytoscape. Visit Aileen Lee’s poster to learn about a new project using
Metaboseek features a convenient interface to run the xcms² pack-
*
333.0608
PTModseek in the Freeman lab.
Fold change
age for MS data processing and feature detection, and tools to filter for
152.9964
the most relevant molecular features using statistical analysis, predic-
350.2119
tion of molecular formulas, annotation of MS2 spectra, MS2 molecular
networking and finding of stable isotope labeled molecules.
0
424.2497
Get Metaboseek
100 200 300 400 500 Metaboseek is available as an installer for Win-
etabo dows, or as a cross-platform R package. You can
Summary Plots download the software and try an online preview
seek OH OH Aggregate data from the feature table into customizable version on our website at
plots for overview and easily create high quality figures.
MS2 Patterns Molecular Formulas
metaboseek.com
O
OH
m/z
Preliminary identifi- C H N O P S ... 350
+ cations for further OH
References Support
analysis R
300
OH
O 10
hacl-1 fold over WT
O
250 1. M. J. Helf, et al., Nat. Commun., 2022, 13, 1.
OH OH O
11:0 HO 2. C. A. Smith, et al., Anal. Chem., 2006, 78, 779.
10:0 N R 1 DFG Fellowship,
200
9:0 H Project #386228702
8:0 3. K. Dührkop, et al., Nat. Methods, 2019, 16, 299.
LC/ HRMS RT (sec) 600 800 1000
4. M. J. Helf, et al., ChemBioChem, 2017, 18, 444.
NIH U01
You might also like
- Manual - Cube Sugar - V02 - ENCNFRDEJP - 190104 PDFDocument36 pagesManual - Cube Sugar - V02 - ENCNFRDEJP - 190104 PDFIgnacio Pérez-Victoria100% (1)
- Asta Analytical MethodsDocument4 pagesAsta Analytical Methodsravivarmahyd8173No ratings yet
- Rivaroxaban Tablets Pharmeuropa 31.02EDocument4 pagesRivaroxaban Tablets Pharmeuropa 31.02EMariaNo ratings yet
- Unit VI Molecular Spectroscopy: Physical ChemistryDocument17 pagesUnit VI Molecular Spectroscopy: Physical ChemistryDewal Deshmukh100% (1)
- Hall of Fame: David Russell Recordings and EditionsDocument5 pagesHall of Fame: David Russell Recordings and EditionsIgnacio Pérez-Victoria100% (1)
- Modelbase System: A Distributed Model Database On The InternetDocument5 pagesModelbase System: A Distributed Model Database On The InternetKirupa KaranNo ratings yet
- Image Retrieval by ExamplesDocument15 pagesImage Retrieval by ExamplesHkh HkhNo ratings yet
- 0 MetaboAnalyst 5.0 OverviewDocument7 pages0 MetaboAnalyst 5.0 Overview유승하No ratings yet
- Performance and Analyses Using Two ETL Extraction Software SolutionsDocument4 pagesPerformance and Analyses Using Two ETL Extraction Software SolutionsVanya SinghalNo ratings yet
- SumitKumarDam - 09-05-2024Document21 pagesSumitKumarDam - 09-05-2024SUMITKUMARDAM[학생](대학원 인공지능학과) No ratings yet
- Entity Relationship Model: Pamantasan NG CabuyaoDocument10 pagesEntity Relationship Model: Pamantasan NG CabuyaoBien MedinaNo ratings yet
- Journal PublishersDocument4 pagesJournal Publishersgraphic designerNo ratings yet
- MetaboAnalyst 5.0 - Narrowing The Gap Between Raw Spectra and Func - 2Document11 pagesMetaboAnalyst 5.0 - Narrowing The Gap Between Raw Spectra and Func - 2Iqbal PrawiraNo ratings yet
- Ali ResumeDocument2 pagesAli Resumeaftab28No ratings yet
- © 2008 University of Delaware, All Rights Reserved Find Us On The Web atDocument2 pages© 2008 University of Delaware, All Rights Reserved Find Us On The Web atkannanmech87No ratings yet
- Poster SurfDocument1 pagePoster SurfEmilio FerraraNo ratings yet
- Week 1Document4 pagesWeek 1katrusampathvishal879No ratings yet
- Leveraging Lightweight Semantics For Search ImprovDocument3 pagesLeveraging Lightweight Semantics For Search ImprovJoffre ReynoldsNo ratings yet
- Weka Software ManualaDocument20 pagesWeka Software ManualaAswin Kumar ThanikachalamNo ratings yet
- DBMS PROJECT Overview - Roll No. 07 & 16Document9 pagesDBMS PROJECT Overview - Roll No. 07 & 16parth vermaNo ratings yet
- Automotive Embedded System SyllabusDocument5 pagesAutomotive Embedded System SyllabusgudduNo ratings yet
- Entity Extraction SystemDocument6 pagesEntity Extraction SystemUday SolNo ratings yet
- A Survey On Software Suites For Data Mining, Analytics and Knowledge DiscoveryDocument6 pagesA Survey On Software Suites For Data Mining, Analytics and Knowledge DiscoveryInternational Journal of computational Engineering research (IJCER)No ratings yet
- Symmetry: Webshell Attack Detection Based On A Deep Super LearnerDocument16 pagesSymmetry: Webshell Attack Detection Based On A Deep Super LearnerDevil izalNo ratings yet
- RE ProposalDocument25 pagesRE ProposalJiun sheng WONGNo ratings yet
- Semantic Product SearchDocument4 pagesSemantic Product SearchYaron RianyNo ratings yet
- CrossMedia: E-Science Collaboration Platform For Effective Multimedia ResearchDocument5 pagesCrossMedia: E-Science Collaboration Platform For Effective Multimedia ResearchAndras MicsikNo ratings yet
- A Modular Reconfigurable and Portable Framework For On-Board Data Processing Architecture and ApplicationsDocument6 pagesA Modular Reconfigurable and Portable Framework For On-Board Data Processing Architecture and ApplicationsBoul chandra GaraiNo ratings yet
- PolyAnalyst Overview ENGDocument31 pagesPolyAnalyst Overview ENGEugene SolomatinNo ratings yet
- Mol ViewerDocument7 pagesMol Viewerwonderopokuasante9No ratings yet
- Eisen PytorchDocument8 pagesEisen PytorchLuboš ŘehounekNo ratings yet
- 1 s2.0 S0169260721004880 MainDocument13 pages1 s2.0 S0169260721004880 MainmaketoNo ratings yet
- Information ExtractionDocument7 pagesInformation ExtractionBini Teflon AnkhNo ratings yet
- Sqmtools: Automated Processing and Visual Analysis of 'Omics Data With R and Anvi'ODocument11 pagesSqmtools: Automated Processing and Visual Analysis of 'Omics Data With R and Anvi'OshanmugapriyaNo ratings yet
- INCOSE IW09-MBSE - Initiative - SysMO - (d01FEB09-aRH-v1.4-sFINAL)Document42 pagesINCOSE IW09-MBSE - Initiative - SysMO - (d01FEB09-aRH-v1.4-sFINAL)Ralph HodgsonNo ratings yet
- Elizabeth Walkup, MacMalwareDocument5 pagesElizabeth Walkup, MacMalwareselavilizationNo ratings yet
- DB MinerDocument6 pagesDB MinerNishant KumarNo ratings yet
- Computers OverviewDocument3 pagesComputers Overviewfiw ahimNo ratings yet
- Patterns For Model-Driven Software-DevelopmentDocument50 pagesPatterns For Model-Driven Software-DevelopmentOtto F OttONo ratings yet
- Professional Program in Data Science and Machine Learning: - ML EngineerDocument4 pagesProfessional Program in Data Science and Machine Learning: - ML EngineerMANPREET SODHINo ratings yet
- Information ExtractionDocument8 pagesInformation ExtractionBini Teflon AnkhNo ratings yet
- Information ExtractionDocument8 pagesInformation ExtractionBini Teflon AnkhNo ratings yet
- Metaseek: A Content-Based Meta-Search Engine For ImagesDocument11 pagesMetaseek: A Content-Based Meta-Search Engine For ImagesEman AlkurdiNo ratings yet
- Dynamic Filtering and Prioritization of Static Code Analysis AlertsDocument2 pagesDynamic Filtering and Prioritization of Static Code Analysis Alertscagla.cengizNo ratings yet
- SIRIUS 4 - A Rapid Tool For Turning Tandem Mass Spectra Into Metabolite Structure InformationDocument10 pagesSIRIUS 4 - A Rapid Tool For Turning Tandem Mass Spectra Into Metabolite Structure InformationJovanderson JacksonNo ratings yet
- Imperial College London (MMG, Yg, Ac901) @doc - Ic.Ac - Uk Inforsense LTDDocument4 pagesImperial College London (MMG, Yg, Ac901) @doc - Ic.Ac - Uk Inforsense LTDNhek VibolNo ratings yet
- General Katalog Ditek JayaDocument52 pagesGeneral Katalog Ditek JayamedisoniantoNo ratings yet
- Information Systems Review of Literatures: II. System ArchitectureDocument3 pagesInformation Systems Review of Literatures: II. System ArchitectureLucman AbdulrachmanNo ratings yet
- Plantweb Optics Data Lake ComponentsDocument2 pagesPlantweb Optics Data Lake ComponentskEWQ 865kNo ratings yet
- Database Management System SKILLING PROJECT-Mojes Film Makers Plan of Action SNO Component Accomplishment Status RemarkDocument1 pageDatabase Management System SKILLING PROJECT-Mojes Film Makers Plan of Action SNO Component Accomplishment Status Remarkprudhvi chNo ratings yet
- Cdisc 2011 Dandamudi PDFDocument1 pageCdisc 2011 Dandamudi PDFKrishna DandamudiNo ratings yet
- Kim 2018Document18 pagesKim 2018Julian Rolando Avendaño PeñaNo ratings yet
- Knowledge Map Design of Bioethics Supporting 3D VisualizationDocument5 pagesKnowledge Map Design of Bioethics Supporting 3D VisualizationStefanyNo ratings yet
- Advance Excel COURSEDocument6 pagesAdvance Excel COURSEjahanviNo ratings yet
- Computer Physics Communications: Vei Wang, Nan Xu, Jin-Cheng Liu, Gang Tang, Wen-Tong GengDocument19 pagesComputer Physics Communications: Vei Wang, Nan Xu, Jin-Cheng Liu, Gang Tang, Wen-Tong Genglixiang RaoNo ratings yet
- Assignment No. 7: Shri Guru Gobind Singhji Institute of Engineering and Technology (SGGSIET), NandedDocument3 pagesAssignment No. 7: Shri Guru Gobind Singhji Institute of Engineering and Technology (SGGSIET), NandedAjay JadhavNo ratings yet
- (IJCST-V12I1P6) :kaushik Kashyap, Rinku Moni Borah, Priyanku Rahang, DR Bornali Gogoi, Prof. Nelson R VarteDocument5 pages(IJCST-V12I1P6) :kaushik Kashyap, Rinku Moni Borah, Priyanku Rahang, DR Bornali Gogoi, Prof. Nelson R VarteEighthSenseGroupNo ratings yet
- Data Warehouse Operational Architecture: Keywords: DB2, Oracle, Data Warehouse Data Warehouse EnvironmentDocument14 pagesData Warehouse Operational Architecture: Keywords: DB2, Oracle, Data Warehouse Data Warehouse EnvironmentRoberto GavidiaNo ratings yet
- Inputs To System DesignDocument8 pagesInputs To System Design555bsyadav555No ratings yet
- Efficient Structure-Informed Featurization and Property Prediction of Ordered, Dilute, and Random Atomic StructuresDocument23 pagesEfficient Structure-Informed Featurization and Property Prediction of Ordered, Dilute, and Random Atomic Structuresdemoc29381hkgsc12343No ratings yet
- ML Training PDFDocument6 pagesML Training PDFshrestha3902No ratings yet
- Conclusion: So Store Manager: It Stores The Sos, Sos MetadataDocument1 pageConclusion: So Store Manager: It Stores The Sos, Sos MetadataSrinivasNo ratings yet
- NASA Constellation Program Ontologies Ralph Hodgson 20080320Document48 pagesNASA Constellation Program Ontologies Ralph Hodgson 20080320Ralph HodgsonNo ratings yet
- UCS RM Intro-EtlDocument30 pagesUCS RM Intro-Etlnur ashfaralianaNo ratings yet
- DATA MINING and MACHINE LEARNING. PREDICTIVE TECHNIQUES: ENSEMBLE METHODS, BOOSTING, BAGGING, RANDOM FOREST, DECISION TREES and REGRESSION TREES.: Examples with MATLABFrom EverandDATA MINING and MACHINE LEARNING. PREDICTIVE TECHNIQUES: ENSEMBLE METHODS, BOOSTING, BAGGING, RANDOM FOREST, DECISION TREES and REGRESSION TREES.: Examples with MATLABNo ratings yet
- Cord-Based Microfluidic Chips As A Platform For ELISA and Glucose AssaysDocument11 pagesCord-Based Microfluidic Chips As A Platform For ELISA and Glucose AssaysIgnacio Pérez-VictoriaNo ratings yet
- Specs PalosDocument3 pagesSpecs PalosIgnacio Pérez-VictoriaNo ratings yet
- Romance D'amour: Arranged Dang Thao NguyenDocument6 pagesRomance D'amour: Arranged Dang Thao NguyenIgnacio Pérez-VictoriaNo ratings yet
- Derive Lab Manual-1Document248 pagesDerive Lab Manual-1Ignacio Pérez-VictoriaNo ratings yet
- Marine Drugs: New 1,4-Dienonesteroids From The Octocoral Dendronephthya SPDocument8 pagesMarine Drugs: New 1,4-Dienonesteroids From The Octocoral Dendronephthya SPIgnacio Pérez-VictoriaNo ratings yet
- Smash 2019 ProgramDocument225 pagesSmash 2019 ProgramIgnacio Pérez-VictoriaNo ratings yet
- Marinedrugs 17 00457Document17 pagesMarinedrugs 17 00457Ignacio Pérez-VictoriaNo ratings yet
- Fender Blacktop & Modern Player ElectricsDocument5 pagesFender Blacktop & Modern Player ElectricsIgnacio Pérez-VictoriaNo ratings yet
- CHAPTER 1-Reading Music and Tablature (TAB) : Intermediate Acoustic GuitarDocument6 pagesCHAPTER 1-Reading Music and Tablature (TAB) : Intermediate Acoustic GuitarIgnacio Pérez-VictoriaNo ratings yet
- Metabolites: The Metarbolomics Toolbox in Bioconductor and BeyondDocument55 pagesMetabolites: The Metarbolomics Toolbox in Bioconductor and BeyondIgnacio Pérez-VictoriaNo ratings yet
- Marine Drugs: Bioactive Bromotyrosine-Derived Alkaloids From The Polynesian Sponge Suberea IanthelliformisDocument16 pagesMarine Drugs: Bioactive Bromotyrosine-Derived Alkaloids From The Polynesian Sponge Suberea IanthelliformisIgnacio Pérez-VictoriaNo ratings yet
- Johann Sebastian BachDocument6 pagesJohann Sebastian BachIgnacio Pérez-VictoriaNo ratings yet
- Metabolites 09 00165 v2Document15 pagesMetabolites 09 00165 v2Ignacio Pérez-VictoriaNo ratings yet
- Marine Drugs: Synthesis and Antitumor Activity Evaluation of Compounds Based On ToluquinolDocument16 pagesMarine Drugs: Synthesis and Antitumor Activity Evaluation of Compounds Based On ToluquinolIgnacio Pérez-VictoriaNo ratings yet
- Marinedrugs 17 00402Document16 pagesMarinedrugs 17 00402Ignacio Pérez-VictoriaNo ratings yet
- Molecules: A Self-Healing and Shape Memory Polymer That Functions at Body TemperatureDocument12 pagesMolecules: A Self-Healing and Shape Memory Polymer That Functions at Body TemperatureIgnacio Pérez-VictoriaNo ratings yet
- Article MoleculesDocument11 pagesArticle MoleculesIgnacio Pérez-VictoriaNo ratings yet
- Marinedrugs 17 00167Document10 pagesMarinedrugs 17 00167Ignacio Pérez-VictoriaNo ratings yet
- Molecules 24 03221Document12 pagesMolecules 24 03221Ignacio Pérez-VictoriaNo ratings yet
- Article MetabolitesDocument25 pagesArticle MetabolitesIgnacio Pérez-VictoriaNo ratings yet
- Beethoven: Selected Works Transcribed For GuitarDocument6 pagesBeethoven: Selected Works Transcribed For GuitarIgnacio Pérez-VictoriaNo ratings yet
- A Systematic Review of Recently Reported Marine Derived Natural Product Kinase InhibitorsDocument38 pagesA Systematic Review of Recently Reported Marine Derived Natural Product Kinase InhibitorsIgnacio Pérez-VictoriaNo ratings yet
- James Ashary Habibie - 191501071 - Jurnal Kafein (ACC NILAI)Document72 pagesJames Ashary Habibie - 191501071 - Jurnal Kafein (ACC NILAI)Habibie TanjungNo ratings yet
- Diffraction GratingDocument9 pagesDiffraction GratingRajaswi BeleNo ratings yet
- LC FinalDocument11 pagesLC FinalCharlez UmerezNo ratings yet
- 1.HPLC Presentation MITDocument58 pages1.HPLC Presentation MITDuc Nhon LENo ratings yet
- Thin Layer ChromatographyDocument8 pagesThin Layer ChromatographyIsabel RinconNo ratings yet
- UV Vis Spectrophotometry Selection GuideDocument9 pagesUV Vis Spectrophotometry Selection GuideDolphingNo ratings yet
- Chapter 1 - Introduction To Spectrometric MethodsDocument57 pagesChapter 1 - Introduction To Spectrometric MethodsFarvin FleetNo ratings yet
- Atomic Absorption SpectrometryDocument64 pagesAtomic Absorption Spectrometryanilrockzzz786No ratings yet
- Forensic Sci Bibliography, ExplosivesDocument4 pagesForensic Sci Bibliography, ExplosivesSusan ColemanNo ratings yet
- Protocolo Purificacion Desde GelDocument3 pagesProtocolo Purificacion Desde GelAriel ArayaNo ratings yet
- Bio 120 Lab Ex 4 PostlabDocument2 pagesBio 120 Lab Ex 4 PostlabSunflowerNo ratings yet
- PRECIPITATIONDocument24 pagesPRECIPITATIONkekoacybelleNo ratings yet
- FTIR ReportDocument15 pagesFTIR Reportapi-3813659100% (10)
- PreviewpdfDocument68 pagesPreviewpdfJoel Ccallo HuaquistoNo ratings yet
- AguerDocument23 pagesAguerHuruk BrosnanNo ratings yet
- Lab.7 عقاقير ثانيDocument9 pagesLab.7 عقاقير ثانيهاني عقيل حسين جوادNo ratings yet
- Spectrochemical Analysis by Atomic Absorption and Emission - Lajunen 2nd Edition 2004Document360 pagesSpectrochemical Analysis by Atomic Absorption and Emission - Lajunen 2nd Edition 2004Goretti Arvizu100% (1)
- Assignment On HPLC and TLC - PDF - Pharmacognosy & PhytochemistryDocument9 pagesAssignment On HPLC and TLC - PDF - Pharmacognosy & PhytochemistryMr HotmasterNo ratings yet
- Lista Peças 2Document5 pagesLista Peças 2mardonio andradeNo ratings yet
- Gatorade Beer's Law Lab-Chem 4-1Document5 pagesGatorade Beer's Law Lab-Chem 4-1Mark Cliffton BadlonNo ratings yet
- Simplified Enrichment Method Using NeedlEx. Application Note (Shimadzu)Document2 pagesSimplified Enrichment Method Using NeedlEx. Application Note (Shimadzu)Maikel Perez NavarroNo ratings yet
- Analytical Techniques in Pharmaceutical Analysis ADocument14 pagesAnalytical Techniques in Pharmaceutical Analysis AKeshavVashistha100% (1)
- Gel Filtration Selection GuideDocument1 pageGel Filtration Selection GuideDolphingNo ratings yet
- Affinity of LDH LabDocument5 pagesAffinity of LDH LabZoe A GarwoodNo ratings yet
- Combined Spectra ProblemsDocument11 pagesCombined Spectra ProblemsUsama khanNo ratings yet
- ChromatographyDocument28 pagesChromatographyyashaviNo ratings yet
- Protein Analysis WorkflowDocument37 pagesProtein Analysis WorkflowKurdianto MSiNo ratings yet