Professional Documents
Culture Documents
AGNES and SPECTRAL CLUSTERING IN R PDF
AGNES and SPECTRAL CLUSTERING IN R PDF
Uploaded by
Sahas ParabOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
AGNES and SPECTRAL CLUSTERING IN R PDF
AGNES and SPECTRAL CLUSTERING IN R PDF
Uploaded by
Sahas ParabCopyright:
Available Formats
AGNES and SPECTRAL CLUSTERING
IN R
Sahas
2023-24-18
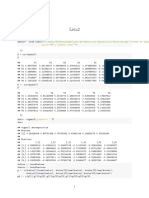
library(tidyverse) #data manipulation
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.1 ✔ readr 2.1.4
## ✔ forcats 1.0.0 ✔ stringr 1.5.0
## ✔ ggplot2 3.4.2 ✔ tibble 3.2.1
## ✔ lubridate 1.9.2 ✔ tidyr 1.3.0
## ✔ purrr 1.0.1
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to be
come errors
library(cluster) #clustering algorithm
library(factoextra) #clustering visualization
## Welcome! Want to learn more? See two factoextra-related books at https://goo.gl/ve3WBa
df=iris
#Complete linkage
hcal=agnes(df,method="complete")
pltree(hcal,cex=0.6, hang= -1, main="Dendrogram of Agnes") # Dendrogram
#Cut into 3 groups
dvcut=cutree(as.hclust(hcal),k=3)
table(dvcut)
## dvcut
## 1 2 3
## 50 66 34
#Single linkage
hcal2=agnes(df,method = "single")
pltree(hcal2,cex=0.6, hang= -1, main="Dendrogram of Agnes") # Dendrogram
#Cut into 3 groups
dvcut=cutree(as.hclust(hcal2), k=3)
table(dvcut)
## dvcut
## 1 2 3
## 50 50 50
#Single linkage
hcal3=agnes(df,method="average")
pltree(hcal3,cex=0.6, hang= -1, main="Dendrogram of Agnes") # Dendrogram
#Cut into 3 groups
dvcut=cutree(as.hclust(hcal3), k=3)
table(dvcut)
## dvcut
## 1 2 3
## 50 51 49
#Spectral Clustering
library(igraph)
##
## Attaching package: 'igraph'
## The following objects are masked from 'package:lubridate':
##
## %--%, union
## The following objects are masked from 'package:dplyr':
##
## as_data_frame, groups, union
## The following objects are masked from 'package:purrr':
##
## compose, simplify
## The following object is masked from 'package:tidyr':
##
## crossing
## The following object is masked from 'package:tibble':
##
## as_data_frame
## The following objects are masked from 'package:stats':
##
## decompose, spectrum
## The following object is masked from 'package:base':
##
## union
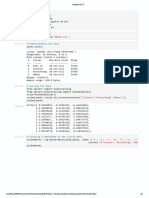
A=rbind(c(0,0.8,0.6,0,0.1,0),c(0.8,0,0.8,0,0,0),c(0.6,0.8,0,0.2,0,0),c(0,0,0.2,0,0.8,0.7),c
(0.1,
0,0,0.8,0,0.8),c(0,0,0,0.7,0.8,0))
A
## [,1] [,2] [,3] [,4] [,5] [,6]
## [1,] 0.0 0.8 0.6 0.0 0.1 0.0
## [2,] 0.8 0.0 0.8 0.0 0.0 0.0
## [3,] 0.6 0.8 0.0 0.2 0.0 0.0
## [4,] 0.0 0.0 0.2 0.0 0.8 0.7
## [5,] 0.1 0.0 0.0 0.8 0.0 0.8
## [6,] 0.0 0.0 0.0 0.7 0.8 0.0
ig= graph.adjacency(A, mode="undirected", weighted = TRUE)
plot(ig)
D= diag(apply(A, 1, sum))
D
## [,1] [,2] [,3] [,4] [,5] [,6]
## [1,] 1.5 0.0 0.0 0.0 0.0 0.0
## [2,] 0.0 1.6 0.0 0.0 0.0 0.0
## [3,] 0.0 0.0 1.6 0.0 0.0 0.0
## [4,] 0.0 0.0 0.0 1.7 0.0 0.0
## [5,] 0.0 0.0 0.0 0.0 1.7 0.0
## [6,] 0.0 0.0 0.0 0.0 0.0 1.5
L=D-A
L
## [,1] [,2] [,3] [,4] [,5] [,6]
## [1,] 1.5 -0.8 -0.6 0.0 -0.1 0.0
## [2,] -0.8 1.6 -0.8 0.0 0.0 0.0
## [3,] -0.6 -0.8 1.6 -0.2 0.0 0.0
## [4,] 0.0 0.0 -0.2 1.7 -0.8 -0.7
## [5,] -0.1 0.0 0.0 -0.8 1.7 -0.8
## [6,] 0.0 0.0 0.0 -0.7 -0.8 1.5
#Eigen values of Laplacian matrix
eig_L= eigen(L)
eig_L$values;
## [1] 2.573487e+00 2.469025e+00 2.285298e+00 2.084006e+00 1.881842e-01
## [6] 2.220446e-15
plot(eig_L$values)
eig_L$vectors
## [,1] [,2] [,3] [,4] [,5] [,6]
## [1,] -0.10598786 0.3788245748 -0.30546656 0.64690880 0.4084006 -0.4082483
## [2,] -0.21517718 -0.7063206485 0.30450981 -0.01441501 0.4418249 -0.4082483
## [3,] 0.36782805 0.3884382495 0.04461661 -0.63818761 0.3713186 -0.4082483
## [4,] -0.61170644 0.0009962363 -0.45451793 -0.33863293 -0.3713338 -0.4082483
## [5,] 0.65221488 -0.3509689196 -0.30495543 0.16645901 -0.4050475 -0.4082483
## [6,] -0.08717145 0.2890305075 0.71581349 0.17786774 -0.4451628 -0.4082483
k=2
Z= eig_L$vectors[,5:6]
Z
## [,1] [,2]
## [1,] 0.4084006 -0.4082483
## [2,] 0.4418249 -0.4082483
## [3,] 0.3713186 -0.4082483
## [4,] -0.3713338 -0.4082483
## [5,] -0.4050475 -0.4082483
## [6,] -0.4451628 -0.4082483
Z[,c(1,2)]=Z[,c(2,1)]
Z
## [,1] [,2]
## [1,] -0.4082483 0.4084006
## [2,] -0.4082483 0.4418249
## [3,] -0.4082483 0.3713186
## [4,] -0.4082483 -0.3713338
## [5,] -0.4082483 -0.4050475
## [6,] -0.4082483 -0.4451628
#Now for the normalization
denom= sqrt(apply(Z^2, 1, sum)) #denominator
denom
## [1] 0.5774580 0.6015612 0.5518552 0.5518654 0.5750914 0.6040171
#denominator2: convert to a matrix
denom2<-replicate(n=k, expr = denom)
denom2
## [,1] [,2]
## [1,] 0.5774580 0.5774580
## [2,] 0.6015612 0.6015612
## [3,] 0.5518552 0.5518552
## [4,] 0.5518654 0.5518654
## [5,] 0.5750914 0.5750914
## [6,] 0.6040171 0.6040171
#Create the Y matrix
y<- Z/denom2
y
## [,1] [,2]
## [1,] -0.7069749 0.7072386
## [2,] -0.6786479 0.7344637
## [3,] -0.7397743 0.6728551
## [4,] -0.7397606 -0.6728702
## [5,] -0.7098842 -0.7043185
## [6,] -0.6758887 -0.7370037
plot(y)
#Apply k-means on y:
spec_clusters<- kmeans(y, centers = 2, nstart=100)
spec_clusters$cluster
## [1] 2 2 2 1 1 1
spec_clusters$size
## [1] 3 3
spec_clusters$centers
## [,1] [,2]
## 1 -0.7085111 -0.7047308
## 2 -0.7084657 0.7048525
You might also like
- 1.1 Read The Data and Do Exploratory Data Analysis. Describe The Data BrieflyDocument50 pages1.1 Read The Data and Do Exploratory Data Analysis. Describe The Data BrieflyP Venkata Krishna Rao100% (19)
- Knn1 MinMaxScalarDocument13 pagesKnn1 MinMaxScalarJoe1No ratings yet
- Import As: PrintDocument4 pagesImport As: PrintI.A.P. JABH RIDETNo ratings yet
- 418 MaterialDocument16 pages418 MaterialiwunzeoziomaNo ratings yet
- Practical 2 51Document5 pagesPractical 2 51Royal EmpireNo ratings yet
- Import As: Numpy NPDocument5 pagesImport As: Numpy NPGilang SaputraNo ratings yet
- Data Mining PortfolioDocument19 pagesData Mining PortfolioJohnMaynardNo ratings yet
- TH C Hành ch4bt3 - SBTDocument10 pagesTH C Hành ch4bt3 - SBTNguyễn OanhNo ratings yet
- Customer Churn in TelecomDocument9 pagesCustomer Churn in TelecomSubrahmanyam JonnalagaddaNo ratings yet
- Lista 2 MVDocument2 pagesLista 2 MVTacio CavalcanteNo ratings yet
- 1 Reducing Dimensionality From 4D To 1D: Numpy NP Pandas PD Sklearn Sklearn - PreprocessingDocument4 pages1 Reducing Dimensionality From 4D To 1D: Numpy NP Pandas PD Sklearn Sklearn - PreprocessingAsmar HajizadaNo ratings yet
- Sales Forecast: October 9, 2020Document9 pagesSales Forecast: October 9, 2020salnasuNo ratings yet
- Heart: Our "Goal" Predict The Presence of Heart Disease in The PatientDocument73 pagesHeart: Our "Goal" Predict The Presence of Heart Disease in The Patientaditya b100% (1)
- Chapter 6. Product Limit Stimator Peter Smith: Required LibrariesDocument9 pagesChapter 6. Product Limit Stimator Peter Smith: Required LibrariesYosef GUEVARA SALAMANCANo ratings yet
- PROYECTO EstadisticaDocument24 pagesPROYECTO EstadisticaBryan MedinaNo ratings yet
- Ditk PPDocument24 pagesDitk PPYna ForondaNo ratings yet
- Import AsDocument27 pagesImport AsFozia Dawood100% (1)
- Recommendation SystemDocument37 pagesRecommendation Systemgsharma_225690No ratings yet
- Sae P5 KautsarDocument13 pagesSae P5 KautsarKepinNo ratings yet
- Data EntryDocument4 pagesData Entry2147033No ratings yet
- Data Science Comp 2Document13 pagesData Science Comp 2tobiasNo ratings yet
- Preprocessing - Preprocessing Your Data With RDocument23 pagesPreprocessing - Preprocessing Your Data With RTonyNo ratings yet
- 20BCE1205 Lab8Document13 pages20BCE1205 Lab8SHUBHAM OJHANo ratings yet
- 03 - K Means Clustering On Iris DatasetsDocument4 pages03 - K Means Clustering On Iris DatasetsJohn WickNo ratings yet
- How To Perform SVAR in R: Mohamed Nachid BoussialaDocument11 pagesHow To Perform SVAR in R: Mohamed Nachid BoussialaMohamed DjellouliNo ratings yet
- Lab6 Results 8974917Document4 pagesLab6 Results 8974917Revathy PNo ratings yet
- Kanseei Tahapt 2Document8 pagesKanseei Tahapt 2ryusrianammarNo ratings yet
- Upyter Notebook1Document5 pagesUpyter Notebook1KRISHVIJAY1012No ratings yet
- R Intro STAT5000Document17 pagesR Intro STAT5000vinay kumarNo ratings yet
- K Means ClusteringDocument10 pagesK Means ClusteringWalid Sassi100% (1)
- Practica 9Document24 pagesPractica 92marlenehh2003No ratings yet
- Import As Import As From Import: "Mean Squared Errors: "Document1 pageImport As Import As From Import: "Mean Squared Errors: "ulNo ratings yet
- ML0101EN Clas Logistic Reg Churn Py v1Document13 pagesML0101EN Clas Logistic Reg Churn Py v1banicx100% (1)
- Chap 35Document62 pagesChap 35Trịnh TâmNo ratings yet
- 212011497-4SE5-Kautsar Hilmi Izzuddin Pertemuan 5Document13 pages212011497-4SE5-Kautsar Hilmi Izzuddin Pertemuan 5KepinNo ratings yet
- 3 Confussion Matrix Hasil Modelling OKDocument8 pages3 Confussion Matrix Hasil Modelling OKArman Maulana Muhtar100% (1)
- 1 2-FramDocument8 pages1 2-Framkrishparakh23No ratings yet
- Kautsar Hilmi Izzuddin - Tugas SAE P5Document13 pagesKautsar Hilmi Izzuddin - Tugas SAE P5KepinNo ratings yet
- Jupyter Notebook Yardlenis Sánchez Act2Document1 pageJupyter Notebook Yardlenis Sánchez Act2NASLYNo ratings yet
- Simulacion Ejercicios Con R: 1.2 SintaxisDocument8 pagesSimulacion Ejercicios Con R: 1.2 SintaxisLimbert Miguel Tola CondoriNo ratings yet
- Handout 02Document12 pagesHandout 02maxi maaeezNo ratings yet
- Source Code in PDF FormatDocument15 pagesSource Code in PDF FormatShivam PrajapatiNo ratings yet
- M0519064 - Muhammad Syahru Aulia - 4Document4 pagesM0519064 - Muhammad Syahru Aulia - 4SalimulhadiNo ratings yet
- Lista 4 - Mineração de DadosDocument15 pagesLista 4 - Mineração de DadosGustavo Figueiredo RamosNo ratings yet
- Homework 5Document104 pagesHomework 5James DakotaNo ratings yet
- ML#07Document21 pagesML#07Kiki NhabindeNo ratings yet
- PCA and FA CommandsDocument13 pagesPCA and FA CommandsNguyễn OanhNo ratings yet
- Hints and AnswersDocument13 pagesHints and Answersashishamitav123No ratings yet
- DALab_Part-B_BCU&BUDocument12 pagesDALab_Part-B_BCU&BU8daudio02No ratings yet
- Chapter 10 - Exercise 10Document6 pagesChapter 10 - Exercise 10krisjooniejin tanNo ratings yet
- PandasDocument21 pagesPandasShubham dattatray koteNo ratings yet
- Program 4 Lu Decomposition C ProgrammingDocument13 pagesProgram 4 Lu Decomposition C ProgrammingPraveen DNo ratings yet
- Debarghya Das (Ba-1), 18021141033Document12 pagesDebarghya Das (Ba-1), 18021141033Rocking Heartbroker DebNo ratings yet
- Output DebugDocument16 pagesOutput DebughaiderNo ratings yet
- PCA Code-CheckpointDocument4 pagesPCA Code-CheckpointАурел ПNo ratings yet
- Credit Card DefaultDocument30 pagesCredit Card Defaulternest muhasaNo ratings yet
- Analisis Dinamico Eje XDocument24 pagesAnalisis Dinamico Eje XVICTOR MANUEL PAITAN MENDEZNo ratings yet
- Lecture Material 8Document9 pagesLecture Material 8Ali NaseerNo ratings yet
- Predicting Diamond Price: 2 Step MethodDocument17 pagesPredicting Diamond Price: 2 Step MethodSarajit Poddar100% (1)