Professional Documents
Culture Documents
0 ratings0% found this document useful (0 votes)
10 viewsLibrary
Library
Uploaded by
Sancella OverallThis document shows code for clustering spatial point data based on distance thresholds. It converts point data to a SpatialPointsDataFrame, calculates distances between points, performs hierarchical clustering, defines clusters based on a distance cutoff, adds cluster assignments to the SpatialPointsDataFrame, computes cluster centroids, plots cluster circles around centroids, and plots the points colored by cluster.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
You might also like
- 21bce8374 DV Lab Week-7Document10 pages21bce8374 DV Lab Week-7scheruvu.30No ratings yet
- Cluster AnalysisDocument1 pageCluster Analysisanjali9414No ratings yet
- Program 4Document25 pagesProgram 4api-709999921No ratings yet
- LocationDocument2 pagesLocationMotivated ToGoalNo ratings yet
- DV Assignment-1Document10 pagesDV Assignment-1scheruvu.30No ratings yet
- Prog 7 Graphs and Charts PDFDocument4 pagesProg 7 Graphs and Charts PDFbharath kNo ratings yet
- 23MCA1104 - Exercise - 10 - Hierarchical Clustering - Ipynb - ColabDocument2 pages23MCA1104 - Exercise - 10 - Hierarchical Clustering - Ipynb - ColabPiyush VermaNo ratings yet
- Codes WorkshopDocument13 pagesCodes Workshopiqra mumtazNo ratings yet
- SodapdfDocument1 pageSodapdfNathon MineNo ratings yet
- R Lab ProgramDocument20 pagesR Lab ProgramRadhiyadevi ChinnasamyNo ratings yet
- Algo Lab8Document7 pagesAlgo Lab8tailormayank6953No ratings yet
- ProjectDocument17 pagesProjectmohamed mohsenNo ratings yet
- Enhanced Db-Scan AlgorithmDocument5 pagesEnhanced Db-Scan AlgorithmseventhsensegroupNo ratings yet
- CGM File - Sharonkaur - It2 - 104Document40 pagesCGM File - Sharonkaur - It2 - 104Sharon kaur katyalNo ratings yet
- Aman DA 111Document14 pagesAman DA 111Adarsh yadavNo ratings yet
- Divisive Hierarchical Clustering Using DIANA TechniqueDocument4 pagesDivisive Hierarchical Clustering Using DIANA Techniquemohamed mohsenNo ratings yet
- Pyspark File Commands and TheoryDocument29 pagesPyspark File Commands and Theorykarangole7074No ratings yet
- Week 10Document15 pagesWeek 10Hanumanthu GouthamiNo ratings yet
- 2.3 Aiml RishitDocument7 pages2.3 Aiml Rishitheex.prosNo ratings yet
- DSM 1Document6 pagesDSM 1no0r32200No ratings yet
- CODIGO#Document4 pagesCODIGO#deger treuriNo ratings yet
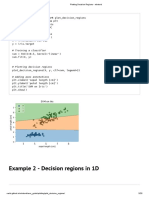
- Plotting Decision Regions - MlxtendDocument5 pagesPlotting Decision Regions - Mlxtendakhi016733No ratings yet
- 19Nh14 102190051 Lab13 Chương Trình MapReduce Shortest Path Using Parallel Breadth First Search BFS 02Document16 pages19Nh14 102190051 Lab13 Chương Trình MapReduce Shortest Path Using Parallel Breadth First Search BFS 02Tri An NguyễnNo ratings yet
- Wa0003Document25 pagesWa0003Ravi MartoliaNo ratings yet
- MessageDocument3 pagesMessageexedrulNo ratings yet
- Machine Learning PractDocument7 pagesMachine Learning PractSunil ShedgeNo ratings yet
- Analisis EspacialesDocument4 pagesAnalisis Espacialesangelica villalob lopezNo ratings yet
- Ğ Message ĞĞĞDocument3 pagesĞ Message ĞĞĞefesisman05No ratings yet
- DSM 2Document7 pagesDSM 2no0r32200No ratings yet
- Data Science LibrariesDocument4 pagesData Science Librariesayushyadav73095No ratings yet
- Lab Distributed Big Data Analytics: Worksheet-3: Spark Graphx and Spark SQL OperationsDocument5 pagesLab Distributed Big Data Analytics: Worksheet-3: Spark Graphx and Spark SQL Operationsbenben08No ratings yet
- HW 1 Math 380 R CodeDocument4 pagesHW 1 Math 380 R CodeTyrome MadkinsNo ratings yet
- Practical-1: Aim:-Write A Program To Implement DDA Line Drawing Algorithm. AlgorithmDocument37 pagesPractical-1: Aim:-Write A Program To Implement DDA Line Drawing Algorithm. AlgorithmDrishti GuptaNo ratings yet
- NearestneighborDocument4 pagesNearestneighborapi-357199306No ratings yet
- 1 - Standard Linear Regression: Numpy NP PandasDocument4 pages1 - Standard Linear Regression: Numpy NP PandasWahib NakhoulNo ratings yet
- Experiment 2.1Document13 pagesExperiment 2.1Utkarsh RastogiNo ratings yet
- Grid Search For SVMDocument9 pagesGrid Search For SVMkPrasad8No ratings yet
- R TutorialDocument3 pagesR TutorialKriti ChopraNo ratings yet
- (Big Data Analytics With PySpark) (CheatSheet)Document7 pages(Big Data Analytics With PySpark) (CheatSheet)Niwahereza DanNo ratings yet
- HARSH CG LAP FileDocument45 pagesHARSH CG LAP FilePritam SharmaNo ratings yet
- CG FIlE DPDocument36 pagesCG FIlE DPNandini GuptaNo ratings yet
- Zulfa Putri Asmawi - TUGAS 10Document8 pagesZulfa Putri Asmawi - TUGAS 10Nurul Isma AsmawiNo ratings yet
- CGIP Lab-FileDocument31 pagesCGIP Lab-FileRahul KumarNo ratings yet
- Main SocketDocument6 pagesMain SocketKiên Lê HữuNo ratings yet
- Dsa Assignment-4 Soutik DeyDocument21 pagesDsa Assignment-4 Soutik DeySoutik DeyNo ratings yet
- Programs: // Program To Implement Scan Conversion of Line Using Direct/Polynomial MethodDocument19 pagesPrograms: // Program To Implement Scan Conversion of Line Using Direct/Polynomial Methodvarun jainNo ratings yet
- Lab7 Hameed 211086Document4 pagesLab7 Hameed 211086Abdul MoaidNo ratings yet
- Liz AssetDocument29 pagesLiz AssetssfofoNo ratings yet
- Codigos RDocument12 pagesCodigos RCarlos Lizárraga RoblesNo ratings yet
- Pair RDD Operations: Flat MapDocument4 pagesPair RDD Operations: Flat Mapmarina duttaNo ratings yet
- Cluster Analysis Hierarchical ClusterDocument12 pagesCluster Analysis Hierarchical Clusterfadhil_ghifariNo ratings yet
- "Computer Graphics & Animation": Practical File ONDocument33 pages"Computer Graphics & Animation": Practical File ONAbhimanyu RanaNo ratings yet
- PGM 7Document3 pagesPGM 7badeniNo ratings yet
- KD Tree DocDocument20 pagesKD Tree Doccs0814No ratings yet
- DSM 3Document6 pagesDSM 3no0r32200No ratings yet
- 新建 文本文档Document6 pages新建 文本文档wangjieNo ratings yet
- Unit2 ML ProgramsDocument7 pagesUnit2 ML Programsdiroja5648No ratings yet
- Spatial Configuration of Epigraphics: What Is An Epigraphic?Document7 pagesSpatial Configuration of Epigraphics: What Is An Epigraphic?Anonymous Ip1ACEhsNo ratings yet
- Experiment No. 1: Graphics Mode InitializationDocument51 pagesExperiment No. 1: Graphics Mode InitializationNitika GuptaNo ratings yet
Library
Library
Uploaded by
Sancella Overall0 ratings0% found this document useful (0 votes)
10 views2 pagesThis document shows code for clustering spatial point data based on distance thresholds. It converts point data to a SpatialPointsDataFrame, calculates distances between points, performs hierarchical clustering, defines clusters based on a distance cutoff, adds cluster assignments to the SpatialPointsDataFrame, computes cluster centroids, plots cluster circles around centroids, and plots the points colored by cluster.
Original Description:
Original Title
library
Copyright
© © All Rights Reserved
Available Formats
PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentThis document shows code for clustering spatial point data based on distance thresholds. It converts point data to a SpatialPointsDataFrame, calculates distances between points, performs hierarchical clustering, defines clusters based on a distance cutoff, adds cluster assignments to the SpatialPointsDataFrame, computes cluster centroids, plots cluster circles around centroids, and plots the points colored by cluster.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
Download as pdf or txt
0 ratings0% found this document useful (0 votes)
10 views2 pagesLibrary
Library
Uploaded by
Sancella OverallThis document shows code for clustering spatial point data based on distance thresholds. It converts point data to a SpatialPointsDataFrame, calculates distances between points, performs hierarchical clustering, defines clusters based on a distance cutoff, adds cluster assignments to the SpatialPointsDataFrame, computes cluster centroids, plots cluster circles around centroids, and plots the points colored by cluster.
Copyright:
© All Rights Reserved
Available Formats
Download as PDF, TXT or read online from Scribd
Download as pdf or txt
You are on page 1of 2
library(sp)
install.packages("sp")
install.packages("rgdall")
install.packages("rgdal")
install.packages("geosphere")
# convert data to a SpatialPointsDataFrame object
xy <- SpatialPointsDataFrame(
matrix(c(x,y), ncol=2), data.frame(ID=seq(1:length(x))),
proj4string=CRS("+proj=longlat +ellps=WGS84 +datum=WGS84"))
# use the distm function to generate a geodesic distance matrix in meters
mdist <- distm(xy)
# cluster all points using a hierarchical clustering approach
hc <- hclust(as.dist(mdist), method="complete")
# define the distance threshold, in this case 40 m
d=40
# define clusters based on a tree "height" cutoff "d" and add them to the SpDataFrame
xy$clust <- cutree(hc, h=d)
install.packages("dismo")
install.packages("rgeos")
library(dismo)
library(rgeos)
# expand the extent of plotting frame
xy@bbox[] <- as.matrix(extend(extent(xy),0.001))
# get the centroid coords for each cluster
cent <- matrix(ncol=2, nrow=max(xy$clust))
for (i in 1:max(xy$clust))
# gCentroid from the rgeos package
cent[i,] <- gCentroid(subset(xy, clust == i))@coords
# compute circles around the centroid coords using a 40m radius
# from the dismo package
ci <- circles(cent, d=d, lonlat=T)
# plot
plot(ci@polygons, axes=T)
plot(xy, col=rainbow(4)[factor(xy$clust)], add=T)
> # define clusters based on a tree "height" cutoff "d" and add them to the SpDataFrame
# define clusters based on a tree "height" cutoff "d" and add them to the SpDataFrame
> xy$clust <- cutree(hc, h=d)
xy$clust <- cutree(hc, h=d)
print xy
print xy&$clust
print(xy&$clust)
print(xy)
xy$clust <- cutree(hc, h=d)
You might also like
- 21bce8374 DV Lab Week-7Document10 pages21bce8374 DV Lab Week-7scheruvu.30No ratings yet
- Cluster AnalysisDocument1 pageCluster Analysisanjali9414No ratings yet
- Program 4Document25 pagesProgram 4api-709999921No ratings yet
- LocationDocument2 pagesLocationMotivated ToGoalNo ratings yet
- DV Assignment-1Document10 pagesDV Assignment-1scheruvu.30No ratings yet
- Prog 7 Graphs and Charts PDFDocument4 pagesProg 7 Graphs and Charts PDFbharath kNo ratings yet
- 23MCA1104 - Exercise - 10 - Hierarchical Clustering - Ipynb - ColabDocument2 pages23MCA1104 - Exercise - 10 - Hierarchical Clustering - Ipynb - ColabPiyush VermaNo ratings yet
- Codes WorkshopDocument13 pagesCodes Workshopiqra mumtazNo ratings yet
- SodapdfDocument1 pageSodapdfNathon MineNo ratings yet
- R Lab ProgramDocument20 pagesR Lab ProgramRadhiyadevi ChinnasamyNo ratings yet
- Algo Lab8Document7 pagesAlgo Lab8tailormayank6953No ratings yet
- ProjectDocument17 pagesProjectmohamed mohsenNo ratings yet
- Enhanced Db-Scan AlgorithmDocument5 pagesEnhanced Db-Scan AlgorithmseventhsensegroupNo ratings yet
- CGM File - Sharonkaur - It2 - 104Document40 pagesCGM File - Sharonkaur - It2 - 104Sharon kaur katyalNo ratings yet
- Aman DA 111Document14 pagesAman DA 111Adarsh yadavNo ratings yet
- Divisive Hierarchical Clustering Using DIANA TechniqueDocument4 pagesDivisive Hierarchical Clustering Using DIANA Techniquemohamed mohsenNo ratings yet
- Pyspark File Commands and TheoryDocument29 pagesPyspark File Commands and Theorykarangole7074No ratings yet
- Week 10Document15 pagesWeek 10Hanumanthu GouthamiNo ratings yet
- 2.3 Aiml RishitDocument7 pages2.3 Aiml Rishitheex.prosNo ratings yet
- DSM 1Document6 pagesDSM 1no0r32200No ratings yet
- CODIGO#Document4 pagesCODIGO#deger treuriNo ratings yet
- Plotting Decision Regions - MlxtendDocument5 pagesPlotting Decision Regions - Mlxtendakhi016733No ratings yet
- 19Nh14 102190051 Lab13 Chương Trình MapReduce Shortest Path Using Parallel Breadth First Search BFS 02Document16 pages19Nh14 102190051 Lab13 Chương Trình MapReduce Shortest Path Using Parallel Breadth First Search BFS 02Tri An NguyễnNo ratings yet
- Wa0003Document25 pagesWa0003Ravi MartoliaNo ratings yet
- MessageDocument3 pagesMessageexedrulNo ratings yet
- Machine Learning PractDocument7 pagesMachine Learning PractSunil ShedgeNo ratings yet
- Analisis EspacialesDocument4 pagesAnalisis Espacialesangelica villalob lopezNo ratings yet
- Ğ Message ĞĞĞDocument3 pagesĞ Message ĞĞĞefesisman05No ratings yet
- DSM 2Document7 pagesDSM 2no0r32200No ratings yet
- Data Science LibrariesDocument4 pagesData Science Librariesayushyadav73095No ratings yet
- Lab Distributed Big Data Analytics: Worksheet-3: Spark Graphx and Spark SQL OperationsDocument5 pagesLab Distributed Big Data Analytics: Worksheet-3: Spark Graphx and Spark SQL Operationsbenben08No ratings yet
- HW 1 Math 380 R CodeDocument4 pagesHW 1 Math 380 R CodeTyrome MadkinsNo ratings yet
- Practical-1: Aim:-Write A Program To Implement DDA Line Drawing Algorithm. AlgorithmDocument37 pagesPractical-1: Aim:-Write A Program To Implement DDA Line Drawing Algorithm. AlgorithmDrishti GuptaNo ratings yet
- NearestneighborDocument4 pagesNearestneighborapi-357199306No ratings yet
- 1 - Standard Linear Regression: Numpy NP PandasDocument4 pages1 - Standard Linear Regression: Numpy NP PandasWahib NakhoulNo ratings yet
- Experiment 2.1Document13 pagesExperiment 2.1Utkarsh RastogiNo ratings yet
- Grid Search For SVMDocument9 pagesGrid Search For SVMkPrasad8No ratings yet
- R TutorialDocument3 pagesR TutorialKriti ChopraNo ratings yet
- (Big Data Analytics With PySpark) (CheatSheet)Document7 pages(Big Data Analytics With PySpark) (CheatSheet)Niwahereza DanNo ratings yet
- HARSH CG LAP FileDocument45 pagesHARSH CG LAP FilePritam SharmaNo ratings yet
- CG FIlE DPDocument36 pagesCG FIlE DPNandini GuptaNo ratings yet
- Zulfa Putri Asmawi - TUGAS 10Document8 pagesZulfa Putri Asmawi - TUGAS 10Nurul Isma AsmawiNo ratings yet
- CGIP Lab-FileDocument31 pagesCGIP Lab-FileRahul KumarNo ratings yet
- Main SocketDocument6 pagesMain SocketKiên Lê HữuNo ratings yet
- Dsa Assignment-4 Soutik DeyDocument21 pagesDsa Assignment-4 Soutik DeySoutik DeyNo ratings yet
- Programs: // Program To Implement Scan Conversion of Line Using Direct/Polynomial MethodDocument19 pagesPrograms: // Program To Implement Scan Conversion of Line Using Direct/Polynomial Methodvarun jainNo ratings yet
- Lab7 Hameed 211086Document4 pagesLab7 Hameed 211086Abdul MoaidNo ratings yet
- Liz AssetDocument29 pagesLiz AssetssfofoNo ratings yet
- Codigos RDocument12 pagesCodigos RCarlos Lizárraga RoblesNo ratings yet
- Pair RDD Operations: Flat MapDocument4 pagesPair RDD Operations: Flat Mapmarina duttaNo ratings yet
- Cluster Analysis Hierarchical ClusterDocument12 pagesCluster Analysis Hierarchical Clusterfadhil_ghifariNo ratings yet
- "Computer Graphics & Animation": Practical File ONDocument33 pages"Computer Graphics & Animation": Practical File ONAbhimanyu RanaNo ratings yet
- PGM 7Document3 pagesPGM 7badeniNo ratings yet
- KD Tree DocDocument20 pagesKD Tree Doccs0814No ratings yet
- DSM 3Document6 pagesDSM 3no0r32200No ratings yet
- 新建 文本文档Document6 pages新建 文本文档wangjieNo ratings yet
- Unit2 ML ProgramsDocument7 pagesUnit2 ML Programsdiroja5648No ratings yet
- Spatial Configuration of Epigraphics: What Is An Epigraphic?Document7 pagesSpatial Configuration of Epigraphics: What Is An Epigraphic?Anonymous Ip1ACEhsNo ratings yet
- Experiment No. 1: Graphics Mode InitializationDocument51 pagesExperiment No. 1: Graphics Mode InitializationNitika GuptaNo ratings yet