Professional Documents
Culture Documents
Bio-Signal Analysis For Smoking
Bio-Signal Analysis For Smoking
Uploaded by
Divya jainOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Bio-Signal Analysis For Smoking
Bio-Signal Analysis For Smoking
Uploaded by
Divya jainCopyright:
Available Formats
In [1]: import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
In [5]: df=pd.read_csv("C:/Users/kisha/OneDrive/Desktop/data sets/smoking.csv")
In [10]: df.drop(columns=["ID","oral"])
df.head()
Out[10]: ID gender age height(cm) weight(kg) waist(cm) eyesight(left) eyesight(right) hearing(left) hearing(rig
0 0 0 40 155 60 81.3 1.2 1.0 1.0
1 1 0 40 160 60 81.0 0.8 0.6 1.0
2 2 1 55 170 60 80.0 0.8 0.8 1.0
3 3 1 40 165 70 88.0 1.5 1.5 1.0
4 4 0 40 155 60 86.0 1.0 1.0 1.0
5 rows × 27 columns
Checking the shape of a dataframe and datatypes of all columns along with calculating the statistical
data.
In [7]: df.shape
(55692, 27)
Out[7]:
In [8]: df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 55692 entries, 0 to 55691
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 55692 non-null int64
1 gender 55692 non-null object
2 age 55692 non-null int64
3 height(cm) 55692 non-null int64
4 weight(kg) 55692 non-null int64
5 waist(cm) 55692 non-null float64
6 eyesight(left) 55692 non-null float64
7 eyesight(right) 55692 non-null float64
8 hearing(left) 55692 non-null float64
9 hearing(right) 55692 non-null float64
10 systolic 55692 non-null float64
11 relaxation 55692 non-null float64
12 fasting blood sugar 55692 non-null float64
13 Cholesterol 55692 non-null float64
14 triglyceride 55692 non-null float64
15 HDL 55692 non-null float64
16 LDL 55692 non-null float64
17 hemoglobin 55692 non-null float64
18 Urine protein 55692 non-null float64
19 serum creatinine 55692 non-null float64
20 AST 55692 non-null float64
21 ALT 55692 non-null float64
22 Gtp 55692 non-null float64
23 oral 55692 non-null object
24 dental caries 55692 non-null int64
25 tartar 55692 non-null object
26 smoking 55692 non-null int64
dtypes: float64(18), int64(6), object(3)
memory usage: 11.5+ MB
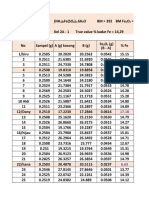
In [10]: df.describe()
Out[10]: ID age height(cm) weight(kg) waist(cm) eyesight(left) eyesight(right)
count 55692.000000 55692.000000 55692.000000 55692.000000 55692.000000 55692.000000 55692.000000
mean 27845.500000 44.182917 164.649321 65.864936 82.046418 1.012623 1.007443
std 16077.039933 12.071418 9.194597 12.820306 9.274223 0.486873 0.485964
min 0.000000 20.000000 130.000000 30.000000 51.000000 0.100000 0.100000
25% 13922.750000 40.000000 160.000000 55.000000 76.000000 0.800000 0.800000
50% 27845.500000 40.000000 165.000000 65.000000 82.000000 1.000000 1.000000
75% 41768.250000 55.000000 170.000000 75.000000 88.000000 1.200000 1.200000
max 55691.000000 85.000000 190.000000 135.000000 129.000000 9.900000 9.900000
8 rows × 24 columns
Checking out the missing values in a dataframe
In [11]: df.isnull().sum()
ID 0
Out[11]:
gender 0
age 0
height(cm) 0
weight(kg) 0
waist(cm) 0
eyesight(left) 0
eyesight(right) 0
hearing(left) 0
hearing(right) 0
systolic 0
relaxation 0
fasting blood sugar 0
Cholesterol 0
triglyceride 0
HDL 0
LDL 0
hemoglobin 0
Urine protein 0
serum creatinine 0
AST 0
ALT 0
Gtp 0
oral 0
dental caries 0
tartar 0
smoking 0
dtype: int64
In [12]: sns.barplot(x=df["gender"],y=df["smoking"])
plt.show()
percentage of the people who are smoking ciggarette.
In [14]: plt.figure(figsize=(10,5))
df["smoking"].value_counts().plot.pie(autopct='%0.2f')
<Axes: ylabel='smoking'>
Out[14]:
In [15]: plt.figure(figsize=(9,6))
sns.histplot(x=df["age"],hue=df["smoking"])
plt.show()
Representation of columns using boxplot to detect outliers. Here outliers represent natural variations in
the population, and they should be left as is in the dataset. These are called true outliers. Therefore for
this dataset we will not remove outliers.
In [21]: for i in df.columns:
if(df[i].dtypes=='int64' or df[i].dtypes=='float64'):
sns.boxplot(df[i])
plt.show()
Data Cleaning
In [41]: from sklearn.preprocessing import LabelEncoder
le=LabelEncoder()
df["smoking"]=le.fit_transform(df["smoking"])
df["gender"]=le.fit_transform(df["gender"])
df["tartar"]=le.fit_transform(df["tartar"])
df["dental caries"]=le.fit_transform(df["dental caries"])
In [15]: df["smoking"].unique()
array([0, 1], dtype=int64)
Out[15]:
Logistic Regression
In [20]: x=df[["gender","height(cm)","Gtp","hemoglobin","triglyceride","age","weight(kg)","wai
y=df["smoking"]
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(x, y, test_size=0.2, random_state= 42)
from sklearn.preprocessing import StandardScaler
sc=StandardScaler()
x_train=sc.fit_transform(x_train)
x_test=sc.transform(x_test)
from sklearn.linear_model import LogisticRegression
lr=LogisticRegression()
lr.fit(x_train,y_train)
y_pred=lr.predict(x_test)
from sklearn.metrics import accuracy_score,classification_report
accuracy_score(y_test,y_pred)
print(classification_report(y_test,y_pred))
precision recall f1-score support
0 0.81 0.76 0.78 7027
1 0.63 0.69 0.66 4112
accuracy 0.73 11139
macro avg 0.72 0.73 0.72 11139
weighted avg 0.74 0.73 0.74 11139
Decision Tree
In [21]: from sklearn.tree import DecisionTreeClassifier
dt= DecisionTreeClassifier()
dt.fit(x_train,y_train)
y_pred=dt.predict(x_test)
print(classification_report(y_test,y_pred))
precision recall f1-score support
0 0.83 0.83 0.83 7027
1 0.70 0.70 0.70 4112
accuracy 0.78 11139
macro avg 0.76 0.76 0.76 11139
weighted avg 0.78 0.78 0.78 11139
Bagging Algorithm – Bagging Classifier
In [32]: from sklearn.ensemble import BaggingClassifier
bagging_clf=BaggingClassifier(base_estimator=DecisionTreeClassifier(),n_estimators=10
bagging_clf.fit(x_train,y_train).score(x_test,y_test)
y_pred=bagging_clf.predict(x_test)
print(classification_report(y_test,y_pred))
C:\Users\kisha\Anaconda\lib\site-packages\sklearn\ensemble\_base.py:166: FutureWarnin
g: `base_estimator` was renamed to `estimator` in version 1.2 and will be removed in
1.4.
warnings.warn(
precision recall f1-score support
0 0.88 0.85 0.86 7027
1 0.75 0.80 0.77 4112
accuracy 0.83 11139
macro avg 0.81 0.82 0.82 11139
weighted avg 0.83 0.83 0.83 11139
Bagging Algorithm-ExtraTrees
In [33]: from sklearn.ensemble import ExtraTreesClassifier
et=ExtraTreesClassifier(n_estimators=1000,random_state=42)
et.fit(x_train,y_train)
y_pred=et.predict(x_test)
print(classification_report(y_test,y_pred))
precision recall f1-score support
0 0.89 0.84 0.86 7027
1 0.75 0.82 0.78 4112
accuracy 0.83 11139
macro avg 0.82 0.83 0.82 11139
weighted avg 0.83 0.83 0.83 11139
Bagging Algorithm -RandomForest
In [34]: from sklearn.ensemble import RandomForestClassifier
rfc=RandomForestClassifier(n_estimators=1000)
rfc.fit(x_train,y_train)
y_pred=rfc.predict(x_test)
print(classification_report(y_test,y_pred))
precision recall f1-score support
0 0.88 0.85 0.86 7027
1 0.75 0.80 0.78 4112
accuracy 0.83 11139
macro avg 0.82 0.83 0.82 11139
weighted avg 0.83 0.83 0.83 11139
In [ ]:
You might also like
- AS Notebook - PCA - Wine Data-4Document1 pageAS Notebook - PCA - Wine Data-4Pranali Malode100% (1)
- Keyword Average Length CurrentlenghtDocument19 pagesKeyword Average Length Currentlenghtjesus salasNo ratings yet
- Experiment 4Document5 pagesExperiment 4Apurva PatilNo ratings yet
- Fashion Golden Package - 50 Merchant - 2022-08-25 14-51-33Document4 pagesFashion Golden Package - 50 Merchant - 2022-08-25 14-51-33Melanie Rizky Hanuransyah -No ratings yet
- Heart Disease Indicator Prediction ModelDocument17 pagesHeart Disease Indicator Prediction ModelSoujanya KerurNo ratings yet
- Logistic RegressionDocument12 pagesLogistic RegressionKagade AjinkyaNo ratings yet
- Counter Strike DataDocument39 pagesCounter Strike DataRahul SharmaNo ratings yet
- Newsvendor TemplateDocument4 pagesNewsvendor TemplateSmit PatelNo ratings yet
- Decision TreeDocument12 pagesDecision TreeKagade AjinkyaNo ratings yet
- Apriori Algo 1Document1 pageApriori Algo 1Us InterpolNo ratings yet
- Assignment 1 Ce627: Structure Design Lab: Satyam Kumar PHD, Roll: 164046010 Indian Institute of Technology, BombayDocument32 pagesAssignment 1 Ce627: Structure Design Lab: Satyam Kumar PHD, Roll: 164046010 Indian Institute of Technology, BombaysatyamNo ratings yet
- BalantaDocument2 pagesBalantaEmil GrigorieNo ratings yet
- Capstone Project 2Document15 pagesCapstone Project 2DavidNo ratings yet
- Cutting OptimizingDocument6 pagesCutting OptimizingPoedya Samapta Catur PamungkasNo ratings yet
- Open Ended LabDocument7 pagesOpen Ended LabZeshan HaidarNo ratings yet
- Diabetes Prediction SystemDocument4 pagesDiabetes Prediction Systemsaurabh khairnarNo ratings yet
- Histogram Equalization and SpecificationDocument1 pageHistogram Equalization and SpecificationLABIDNo ratings yet
- Gopal Ji PakshikiDocument150 pagesGopal Ji PakshikiNIRBHAY PANDEYNo ratings yet
- Loannis K., Example 14.1, Page 896 Rev01Document51 pagesLoannis K., Example 14.1, Page 896 Rev01rafzaltronNo ratings yet
- Healthcare PGPDocument28 pagesHealthcare PGPSneh PrakharNo ratings yet
- Crypto: Name Price ($) Quantity Deposit Fixed Price ($) Fixed Quantity Fixed Profit ($)Document6 pagesCrypto: Name Price ($) Quantity Deposit Fixed Price ($) Fixed Quantity Fixed Profit ($)danyl maslowNo ratings yet
- Olah Data Monica JOGLO REV2Document4 pagesOlah Data Monica JOGLO REV2Lia WieNo ratings yet
- Alphabet - Kaggal - Jupyter NotebookDocument6 pagesAlphabet - Kaggal - Jupyter Notebookankitasonawane9988No ratings yet
- Queueing SystemDocument5 pagesQueueing SystemLucela SisonNo ratings yet
- Evaluacion de La Integral de Duhamel para Cargas SismicasDocument6 pagesEvaluacion de La Integral de Duhamel para Cargas SismicasAngela Odría RubioNo ratings yet
- Solution LabAssignmentDocument15 pagesSolution LabAssignmentHitarth Anand RohraNo ratings yet
- Data Input RockworkDocument14 pagesData Input RockworkRahayanti PrihartiniNo ratings yet
- Sap Adi WiryantaraDocument142 pagesSap Adi WiryantaraAgus ParwataNo ratings yet
- Examples On Comparison of Survival CurvesDocument3 pagesExamples On Comparison of Survival CurvesKimondo KingNo ratings yet
- Ejericios OPE III-IPQ-NAYSU - VENTURA - POCCOHUANCADocument51 pagesEjericios OPE III-IPQ-NAYSU - VENTURA - POCCOHUANCANAYSU VENTURA POCCOHUANCANo ratings yet
- Loading The Dataset: 'Diabetes - CSV'Document4 pagesLoading The Dataset: 'Diabetes - CSV'Divyani ChavanNo ratings yet
- Diagram C Rizka Yuni ZhafiraDocument11 pagesDiagram C Rizka Yuni ZhafiraRizka Yuni ZhafiraNo ratings yet
- Riset OperasionalDocument12 pagesRiset OperasionalAde HendrawanNo ratings yet
- Riset OperasionalDocument12 pagesRiset OperasionalAde HendrawanNo ratings yet
- Logistic Regression 205Document8 pagesLogistic Regression 205Ranadeep DeyNo ratings yet
- Income Qualification Project3Document40 pagesIncome Qualification Project3NikhilNo ratings yet
- Observation: As We Can See We Have Threwe Types of Datatypes I.E. (Int, Float, Object) That Means We Have Both Categorical and Numerical DataDocument2 pagesObservation: As We Can See We Have Threwe Types of Datatypes I.E. (Int, Float, Object) That Means We Have Both Categorical and Numerical DataPRATIK RATHODNo ratings yet
- Correlation and Regression AnalysisDocument14 pagesCorrelation and Regression AnalysisMohammed IjasNo ratings yet
- A5 A.ipynb - ColaboratoryDocument8 pagesA5 A.ipynb - Colaboratoryswetank.raut22No ratings yet
- Output Sap2000 Tugas BajaDocument98 pagesOutput Sap2000 Tugas BajaAgus ParwataNo ratings yet
- Diskusi 4 VIVIDocument2 pagesDiskusi 4 VIVISandiman NurNo ratings yet
- ESCALERA1Document157 pagesESCALERA1David LondoñoNo ratings yet
- Laporan 2015 RumusDocument107 pagesLaporan 2015 RumusPuskesmas AmahaiNo ratings yet
- 40513-PG2 Mise en Route.o7000LKMdbmDocument17 pages40513-PG2 Mise en Route.o7000LKMdbmJustin PathatyNo ratings yet
- Leger Nilai Rapor Kelas KELAS XI MM 1Document4 pagesLeger Nilai Rapor Kelas KELAS XI MM 1Naya Honey BeeNo ratings yet
- Leger Nilai Rapor Kelas KELAS XI MMDocument4 pagesLeger Nilai Rapor Kelas KELAS XI MMNaya Honey BeeNo ratings yet
- Assignment4 VidulGargDocument14 pagesAssignment4 VidulGargvidulgarg1524No ratings yet
- Item Analysis Using CITAS - Demonstration (Empty)Document115 pagesItem Analysis Using CITAS - Demonstration (Empty)Al-John EspejoNo ratings yet
- Triangulo PascalDocument6 pagesTriangulo Pascaljuan carlos ñato yepezNo ratings yet
- Healthcare PGP - Capstone ProjectDocument32 pagesHealthcare PGP - Capstone ProjectSneh Prakhar0% (1)
- Untitled4 Assigment 3Document9 pagesUntitled4 Assigment 3eigintaeeNo ratings yet
- Equipo 1 - Actividad A4U2 - 5SBDocument17 pagesEquipo 1 - Actividad A4U2 - 5SBCLAUDIO DE LA CRUZ SANTOSNo ratings yet
- NHL 11 OptimizerDocument9 pagesNHL 11 OptimizerSGRockerJoshNo ratings yet
- Tabel Hasil Uji Validitas Dan ReliabilitasDocument3 pagesTabel Hasil Uji Validitas Dan Reliabilitasabimanyu wirantoNo ratings yet
- X Xbarr Chart: AverageDocument5 pagesX Xbarr Chart: AverageAniceto Salomão MachavaNo ratings yet
- Data Praktik GravimetriDocument68 pagesData Praktik GravimetriNadhia OctavianiNo ratings yet
- Stock AbhinandanDocument5 pagesStock Abhinandanmyvillagelife2410No ratings yet
- Računanje Cevi 48.3X2Document32 pagesRačunanje Cevi 48.3X2Marko MarkovicNo ratings yet
- SPC 26965 En8dcr Ø65Document1 pageSPC 26965 En8dcr Ø65HAMPI JBNo ratings yet
- The Giant: Numbers 0–20 Activity Book, Ages 4 - 5From EverandThe Giant: Numbers 0–20 Activity Book, Ages 4 - 5No ratings yet
- Impact of Occupant Autonomy On Satisfaction and Building Energy EffciencyDocument9 pagesImpact of Occupant Autonomy On Satisfaction and Building Energy EffciencyAngelica ElboNo ratings yet
- Wire Gauge and Current LimitsDocument3 pagesWire Gauge and Current LimitssreelakshmisnrNo ratings yet
- Shoolini University Mid SemDocument3 pagesShoolini University Mid SemClash ClanNo ratings yet
- Periodicity Review SL KeyDocument4 pagesPeriodicity Review SL KeyYeyoung ParkNo ratings yet
- CephalosporinDocument14 pagesCephalosporinمريم يحيى كاظم يحيىNo ratings yet
- Superacid PresentationDocument14 pagesSuperacid PresentationTushar TanejaNo ratings yet
- No Course Course Description Credit HourDocument3 pagesNo Course Course Description Credit HourfazaseikoNo ratings yet
- Amateur Radio QRP Projects - March 2011Document23 pagesAmateur Radio QRP Projects - March 2011victorplugaruNo ratings yet
- Sample Test Paper: Le Feuv Egùke VadDocument18 pagesSample Test Paper: Le Feuv Egùke VadFarhanNo ratings yet
- CE Overbooking (RAN18.1 - 02)Document96 pagesCE Overbooking (RAN18.1 - 02)Cedric Kouamen DjandaNo ratings yet
- Experimental and Analysis of Welding Electrode Wire Straightening and Cutting MachineDocument14 pagesExperimental and Analysis of Welding Electrode Wire Straightening and Cutting MachineTarig shgool HagarNo ratings yet
- Polymers 11 01543 v2Document11 pagesPolymers 11 01543 v2yazarizcizerizNo ratings yet
- Test 4 BPSC Main Exam 2019: Assistant EngineerDocument9 pagesTest 4 BPSC Main Exam 2019: Assistant EngineerAmit KumarNo ratings yet
- Klocke Kroemer ICG15Document11 pagesKlocke Kroemer ICG15ranim najibNo ratings yet
- EN Wind Code - Part2Document7 pagesEN Wind Code - Part2AJBAJB BAJBAJNo ratings yet
- Seeing and Feeling: A Comparison of Van Gogh and CezanneDocument8 pagesSeeing and Feeling: A Comparison of Van Gogh and CezanneAlli CampbellNo ratings yet
- Nail Making Machine 20-21Document8 pagesNail Making Machine 20-21parhamadkNo ratings yet
- c3d Content Italy 2017 ItalianDocument128 pagesc3d Content Italy 2017 ItaliancamulNo ratings yet
- Wave Optics MCQDocument8 pagesWave Optics MCQGuru ki physicsNo ratings yet
- MM Unit-III - 0Document22 pagesMM Unit-III - 0Sarthak GudwaniNo ratings yet
- Gl90ad PDFDocument2 pagesGl90ad PDFCarlos CornielesNo ratings yet
- Msat-Xee-8 2 - 30 2Document2 pagesMsat-Xee-8 2 - 30 2GingubaNo ratings yet
- Fully Dispersed RH Ensemble Catalyst To Enhance Low-Temperature ActivityDocument23 pagesFully Dispersed RH Ensemble Catalyst To Enhance Low-Temperature ActivityS Tunkla EcharojNo ratings yet
- Question Paper - (Electrical) : Maximum Marks: 50 Time Allowed: 1 HourDocument6 pagesQuestion Paper - (Electrical) : Maximum Marks: 50 Time Allowed: 1 Hourbikas_sahaNo ratings yet
- KF 4 N 20 LWDocument6 pagesKF 4 N 20 LWJosé Manuel Izea NavarroNo ratings yet
- Lect 16 - 17 - Mechanism of Energy TransportDocument9 pagesLect 16 - 17 - Mechanism of Energy TransportSatyajit SamalNo ratings yet
- Activating Generic Object Services (GOS) Toolbar in SAP ObjectsDocument26 pagesActivating Generic Object Services (GOS) Toolbar in SAP Objectsabhishek100% (2)
- Work Energy Chapter Problems-2009-05-13Document21 pagesWork Energy Chapter Problems-2009-05-13Liam ReillyNo ratings yet
- JCL UtilitiesDocument5 pagesJCL UtilitiespallaviNo ratings yet
- Electric Vehicle Drive Simulation With MATLAB/SimulinkDocument24 pagesElectric Vehicle Drive Simulation With MATLAB/SimulinkSingam SridharNo ratings yet