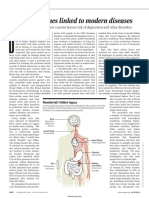
Professional Documents
Culture Documents
Prim Art 3
Prim Art 3
Uploaded by
Tamyy VuOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Prim Art 3
Prim Art 3
Uploaded by
Tamyy VuCopyright:
Available Formats
RES EARCH
Y EVOLUTION of the human Y chromosome (Fig. 1B) (14).
This yielded sequence coverage of 1.4× for
The evolutionary history of Neanderthal Denisova 4, 3.5× for Denisova 8, 0.8× for Spy
94a, and 14.3× for Mezmaiskaya 2 (Fig. 1C and
and Denisovan Y chromosomes table S2). In addition, we used a capture array
designed for modern human Y chromosomes
Martin Petr1*, Mateja Hajdinjak1,2, Qiaomei Fu3,4,5, Elena Essel1, Hélène Rougier6, Isabelle Crevecoeur7, (3) to obtain 7.9× coverage of ~560 kb of the
Patrick Semal8, Liubov V. Golovanova9, Vladimir B. Doronichev9, Carles Lalueza-Fox10, Y chromosome from the ~46- to 53-ka-old El
Marco de la Rasilla11, Antonio Rosas12, Michael V. Shunkov13, Maxim B. Kozlikin13, Sidrón 1253 Neanderthal (Fig. 1C and table S2),
Anatoli P. Derevianko13, Benjamin Vernot1, Matthias Meyer1, Janet Kelso1* which has been analyzed previously (15, 16).
To call genotypes of the captured archaic hu-
Ancient DNA has provided new insights into many aspects of human history. However, we lack man and previously published modern human
comprehensive studies of the Y chromosomes of Denisovans and Neanderthals because the majority of Y chromosomes (4, 17, 18), we leveraged the
specimens that have been sequenced to sufficient coverage are female. Sequencing Y chromosomes from haploid nature of the human Y chromosome
two Denisovans and three Neanderthals shows that the Y chromosomes of Denisovans split around and implemented a consensus approach that
700 thousand years ago from a lineage shared by Neanderthals and modern human Y chromosomes, which requires at least 90% of the reads observed at
diverged from each other around 370 thousand years ago. The phylogenetic relationships of archaic and each site covered by at least three reads to agree
modern human Y chromosomes differ from the population relationships inferred from the autosomal on a single allele (14). This minimizes the im-
genomes and mirror mitochondrial DNA phylogenies, indicating replacement of both the mitochondrial and pact of aDNA damage on genotyping accuracy
Y chromosomal gene pools in late Neanderthals. This replacement is plausible if the low effective population while allowing for a small amount of sequenc-
size of Neanderthals resulted in an increased genetic load in Neanderthals relative to modern humans. ing error or contamination (fig. S8) (14).
To determine the relationships between
A
Denisovan, Neanderthal, and modern human
ncient DNA (aDNA) has transformed our The mtDNA and autosomal sequences of Y chromosomes, we constructed a neighbor-
understanding of human evolutionary Neanderthals, Denisovans, and modern humans joining tree from the Y chromosome geno-
history, revealing complex patterns of have revealed puzzling phylogenetic discrep- type calls (14). Unlike the rest of the nuclear
population migration and gene flow, in- ancies. Autosomal genomes show that Nean- genome, which puts Denisovans and Nean-
cluding admixture from archaic humans derthals and Denisovans are sister groups derthals as sister groups to modern humans (2),
into modern humans. Particularly important that split from modern humans between the Denisovan Y chromosomes form a separate
have been analyses of autosomal sequences 550 thousand and 765 thousand years (ka) ago lineage that split before Neanderthal and mod-
(1, 2), which represent a composite of geneal- (6). By contrast, the mtDNAs of Neanderthals ern human Y chromosomes diverged from each
ogies of any individual’s ancestors. Although and modern humans are more similar to one other (Fig. 2A). Notably, all three late Neander-
mitochondrial DNA (mtDNA) and Y chromo- another [time to the most recent common thal Y chromosomes cluster together and fall
somes only provide information about single ancestor (TMRCA) of 360 to 468 ka ago] than outside of the variation of present-day human
maternal and paternal lineages, they offer a to the mtDNAs of Denisovans (7). Notably, Y chromosomes (Fig. 2A).
distinctive perspective on various aspects of ~400-ka-old early Neanderthals from Sima de To estimate the TMRCA of archaic and mod-
population history such as sex-specific migra- los Huesos were shown to carry mitochon- ern human Y chromosomes, we adapted a
tion, matrilocality and patrilocality, and variance drial genomes related to Denisovan mtDNAs previously published method that calculates
in reproductive success between individuals (8, 9). This suggests that Neanderthals origi- the archaic-modern human TMRCA as a pro-
(3–5). Furthermore, because of their lower ef- nally carried a Denisovan-like mtDNA, which portion of the deepest known split in present-
fective population size (Ne) compared with that was later completely replaced through ancient day human Y variation (4, 10, 14) and is therefore
of autosomal loci, coalescent times of mtDNA gene flow from an early lineage related to robust to low coverage and aDNA damage (10)
and Y chromosomes sampled from two pop- modern humans (7, 9). (fig. S8 and table S2). We first calculated the
ulations provide an upper bound for the last The Y chromosomes of Neanderthals and mutation rate in the 6.9-Mb target region to be
time they experienced gene flow. Denisovans should provide an additional source 7.34 × 10−10 per base pair per year [bootstrap
of information about population splits and confidence interval (CI) 6.27 × 10−10 to 8.46 ×
1
Department of Evolutionary Genetics, Max Planck Institute gene flow events between archaic and mod- 10−10] (fig. S11 and table S11) (14) and used it
for Evolutionary Anthropology, D-04103 Leipzig, Germany. 2The ern humans or populations related to them. to estimate a TMRCA of ~249 ka ago (boot-
Francis Crick Institute, NW1 1AT London, UK. 3Key Laboratory of However, with the exception of a small strap CI 213 to 293 ka ago) (fig. S11 and table
Vertebrate Evolution and Human Origins of Chinese Academy of
Sciences, IVPP, CAS, Beijing 100044, China. 4CAS Center for amount of Neanderthal Y chromosome cod- S11) (14) for the African A00 lineage and a set
Excellence in Life and Paleoenvironment, Beijing 100044, China. ing sequence (118 kb) (10), none of the male of non-African Y chromosomes (4, 18). This is
5
University of Chinese Academy of Sciences, Beijing 100049, Neanderthals or Denisovans studied to date consistent with other studies of present-day
China. 6Department of Anthropology, California State University,
Northridge, Northridge, CA 91330-8244, USA. 7Université de
have yielded sufficient amounts of endoge- human Y chromosomes (4, 17), suggesting that
Bordeaux, CNRS, UMR 5199-PACEA, 33615 Pessac Cedex, nous DNA to allow comprehensive studies of the Y chromosomal regions we sequenced are
France. 8Royal Belgian Institute of Natural Sciences, 1000 archaic human Y chromosomes. not unusual in terms of their mutation rate.
Brussels, Belgium. 9ANO Laboratory of Prehistory 14 Linia 3-11,
St. Petersburg 1990 34, Russia. 10Institute of Evolutionary
Previous genetic studies identified two male We then used this A00 divergence time of
Biology, Consejo Superior de Investigaciones Científicas, Denisovans, Denisova 4 (55 to 84 ka old) and 249 ka ago to infer TMRCAs between archaic
Universitat Pompeu Fabra, 08003 Barcelona, Spain. 11Área de Denisova 8 (106 to 136 ka old) (11, 12), and two Y chromosomes and present-day non-African
Prehistoria, Departamento de Historia, Universidad de Oviedo,
male late Neanderthals, Spy 94a (38 to 39 ka old) Y chromosomes for each archaic individual
33011 Oviedo, Spain. 12Departamento de Paleobiología, Museo
Nacional de Ciencias Naturales, Consejo Superior de and Mezmaiskaya 2 (43 to 45 ka old) (13) (fig. S14 and table S12) (14). The two Denisovan
Investigaciones Científicas, 28006 Madrid, Spain. 13Institute of (Fig. 1A). To enrich for Y chromosome DNA Y chromosomes split from the modern human
Archaeology and Ethnography, Siberian Branch, Russian from these individuals, we performed hybrid- lineage around 700 ka ago (Denisova 8: 707 ka
Academy of Sciences, Novosibirsk, Russia.
*Corresponding author. Email: mp@bodkan.net (M.P.); kelso@ ization capture using probes we designed to ago, CI 607 to 835 ka ago; Denisova 4: 708 ka
eva.mpg.de (J.K.) target ~6.9 Mb of the nonrecombining portion ago, CI 550 to 932 ka ago) (Fig. 2B and table
Petr et al., Science 369, 1653–1656 (2020) 25 September 2020 1 of 4
RES EARCH | REPOR T
Denisova 8
Denisova 4
El Sidrón 1253
Mezmaiskaya 2
Spy 94a
150 125 100 75 50 0 age [ka ago]
B 0 Mb 10 Mb 20 Mb 30 Mb
ampliconic X−degenerate
heterochromatic X−transposed
pseudo−autosomal others
C Denisova 4
20X
10X
2X
20X
Denisova 8 10X
2X
20X
Spy 94a 10X
2X
20X
Mezmaiskaya 2 10X
2X
El Sidrón 1253 20X
10X
(560 kb) 2X
0X 5X 10X 15X 20X 25X
coverage per site
Fig. 1. Overview of male archaic humans in our study. (A) Archaeological site (C) (Left) Spatial distribution of sequencing coverage along the ~6.9 Mb of capture
locations. Ages of specimens are shown as an inset (12, 13, 15). (B) Portion of the target regions. The heights of the thin vertical bars represent average coverage in
human Y chromosome targeted for capture [legend on right, coordinates of genomic each target region. Coordinates are aligned to match the chromosome shown in (B).
regions are from (30)]. Thin black vertical lines show individual target capture regions. (Right) Coverage across all target sites for each individual to the left.
S12). By contrast, the three Neanderthal Y chro- timate was calculated from ~3× coverage of (Fig. 3A) (7). Previous work indicates that the
mosomes split from the modern human lineage 118 kb of nuclear exome capture sequence and, rate of gene flow from modern humans into
about 370 ka ago: 353 ka ago for Spy 94a (CI 287 because of the limited amount of data, used Neanderthals was on the order of only a few
to 450 ka ago), 370 ka ago for Mezmaiskaya 2 single-nucleotide polymorphisms supported percent (20, 21). Because the fixation proba-
(CI 326 to 420 ka ago), and 339 ka ago for even by single reads (10, 16). However, this is bility of a locus is equal to its initial frequency
El Sidrón 1253 (CI 275 to 408 ka ago) (Fig. 2B problematic because it can result in an in- in a population (22), the joint probability of
and table S12). Additionally, we used the pro- creased rate of erroneously called genotypes, both Neanderthal mtDNA and Y chromo-
portion of sharing of derived alleles with the leading to some derived alleles that El Sidrón somes being replaced by their introgressed
high-coverage Mezmaiskaya 2 to estimate the 1253 shares with both Neanderthals and mod- modern human counterparts starting from a
TMRCA of the three Neanderthal Y chromo- ern humans being converted to the ancestral low initial frequency is even lower. However,
somes to around 100 ka ago (figs. S25 and S26). state, increasing the apparent TMRCA. When owing to their low Ne and reduced efficacy of
We validated the robustness of all TMRCA es- we applied filtering designed to mitigate er- purifying selection, Neanderthals have been
timates using filters of varying levels of strin- rors (14) to the original El Sidrón 1253 data, we shown to have accumulated an excess of de-
gency and different genotype calling methods arrived at TMRCA estimates for El Sidrón 1253 leterious variation compared with modern
and also by comparing capture and shotgun consistent with all other Neanderthals in our humans (16), and it has been suggested that
sequence results (figs. S19, S21, and S23). Al- study (fig. S22). introgressed DNA was not neutral (23, 24).
though there was some evidence of capture The Denisovan–modern human Y chromo- To explore the dynamics of modern human
bias in the data (fig. S7), we observed no con- some TMRCA estimates agree with population Y chromosomes introgressed into Neanderthals,
sistent differences between capture data and split times inferred from autosomal sequences, we simulated introgression of a nonrecombin-
shotgun sequences or between individuals suggesting that the differentiation of Denisovan ing, uniparental locus under purifying selec-
showing different read length distributions, Y chromosomes from modern humans occurred tion (14, 25). We considered a range of values
indicating that technical biases do not affect through a simple population split (19). By con- for the following parameters: Neanderthal and
our inferences (fig. S21). trast, the young TMRCA of Neanderthal and modern human Ne, the time that both popula-
Our estimates of the Neanderthal–modern modern human Y chromosomes and mtDNAs tions evolved independently after their split,
human TMRCA (Fig. 2B) are younger than suggest that these loci have been replaced in and the amount of sequence under selection,
the previous estimate of ~588 ka ago from the Neanderthals through gene flow from an early all of which affect the amount of deleterious
El Sidrón 1253 individual (10). This older es- lineage closely related to modern humans variation that accumulated in Neanderthal
Petr et al., Science 369, 1653–1656 (2020) 25 September 2020 2 of 4
RES EARCH | REPOR T
A Denisova 4 B 1,000,000
100
TMRCA with non−Africans [years ago]
Denisova 8
750,000
100 Spy 94a
100
Mezmaiskaya 2
96
500,000
100 El Sidrón 1253
A00
100
non−Africans
250,000
a4 a8 12
53 a2 4a A0
0
ov ov ay y9
e nis e nis d r ón a isk Sp
D D Si zm
El Me
Fig. 2. Phylogenetic relationships between archaic and modern human (B) Estimates of TMRCA between Y chromosomes along the x axis and a panel
Y chromosomes. (A) Neighbor-joining tree estimated from the Y chromosome of 13 non-African Y chromosomes. Each dot represents the TMRCA with a
genotype calls, excluding C-to-T and G-to-A polymorphisms, rooted with a single non-African Y chromosome, with error bars showing 95% CI from a
chimpanzee as the outgroup (14). Numbers show bootstrap support out of resampling of branch counts (14). Black horizontal lines show the mean
100 bootstrap replicates. Terminal branch lengths are not informative about TMRCA calculated across the full non-African panel (dashed lines) with
the ages of specimens (Fig. 1A), owing to differences in sequence quality. resampling-based 95% CI (solid lines) (14).
A 1500
B 1.00
1200 fitness
TMRCA [ka ago]
reduction
900 ~10%
replacement probability
0.75
~9%
600 ~8%
~7%
~6%
300 0.50 ~5%
~4%
~3%
~2%
0.25 ~1%
Modern humans
neutrality
Neanderthals
Denisovans
0.00
0 10000 20000 30000 40000 50000
time after gene flow [years]
Fig. 3. Proposed model for the replacement of Neanderthal Y chromosomes population split time between archaic and modern humans, shown as the
and mtDNA. (A) Relationships between archaic and modern human mtDNA dotted red horizontal line (6). (B) Probability of replacement of a non-
and Y chromosomes. The semitransparent Neanderthal lineage indicates recombining, uniparental Neanderthal locus over time, assuming a given level
a (as yet unsampled) hypothetical Y chromosome replaced by an early lineage of fitness burden relative to its modern human counterpart. Trajectories are
related to modern human Y chromosomes. Most recent common ancestors based on forward simulations across a grid of parameters (figs. S27 to S29) (14),
with modern human lineages are shown for mtDNA (circles) and Y chromosomes with Ne of modern humans and Neanderthals fixed at 10,000 and 1000,
(triangles). The inset shows TMRCAs for the four nodes in the diagram: respectively. Modern human introgression was simulated in a single pulse
Y chromosome TMRCAs as estimated by our study and mtDNA TMRCA estimates at 5%. Replacement probabilities from a wider range of model parameters
from the literature (7, 8). The red shaded area highlights the 95% CI for the are shown in fig. S31.
and modern human populations before in- gressed modern human Y chromosomes in deleterious mutations on each simulated
trogression (14). We simulated introgression Neandertals over 100 ka. For each combina- chromosome (14). This allows us to make
of modern human Y chromosomes into the tion of parameters, we calculated how much a general statement about the probability
Neanderthal population in a single pulse and lower the fitness of an average Neanderthal of replacement in terms of the difference
varied the contribution between 1 and 10%. Y chromosome is compared with an average in fitness between Neanderthal and modern
We then traced the frequency of the intro- modern human Y chromosome using all linked human Y chromosomes while abstracting
Petr et al., Science 369, 1653–1656 (2020) 25 September 2020 3 of 4
RES EARCH | REPOR T
over other factors that affect reproductive fit- groups (10). Furthermore, we predict that the 27. J. W. Ballard, M. C. Whitlock, Mol. Ecol. 13, 729–744
ness but are currently impossible to simulate ~400-ka-old Sima de los Huesos Neanderthals (2004).
28. T. Bonnet, R. Leblois, F. Rousset, P.-A. Crochet, Evolution 71,
accurately (26). should carry a Y chromosome lineage more 2140–2158 (2017).
For example, assuming 5% gene flow from similar to that of Denisovans than to that of 29. F. A. Seixas, P. Boursot, J. Melo-Ferreira, Genome Biol. 19, 91
modern humans, we found that even a 1% re- later Neanderthals (8, 9). Although the amount (2018).
30. L. Skov, M. H. Schierup; Danish Pan Genome Consortium,
duction in Neanderthal Y chromosome fitness of modern human gene flow into Neanderthals PLOS Genet. 13, e1006834 (2017).
increases the probability of replacement after appears to have been limited (13, 20, 21), we 31. M. Petr, “The evolutionary history of Neandertal and Denisovan
50 ka to ~25%, and a 2% reduction in fitness demonstrate that the replacement of mtDNA Y chromosomes” - code and Jupyter notebooks. Zenodo
(2020); doi:10.5281/zenodo.3941654.
increases this probability to ~50% (Fig. 3B). and Y chromosomes in Neanderthals is highly 32. M. Petr, “The evolutionary history of Neandertal and Denisovan
However, the rate of gene flow as well as any plausible, given the higher genetic load in Y chromosomes” - Y chromosome capture designs. Zenodo
factor that contributes to the difference in Neanderthals compared with that in modern (2020); doi: 10.5281/zenodo.3940568.
fitness between Neanderthal and modern hu- humans. Our results imply that differences in
AC KNOWLED GME NTS
man Y chromosomes will have a pronounced genetic load in uniparental loci between two We thank S. Pääbo, M. Stoneking, B. Peter, M. Slatkin, L. Skov,
effect on the replacement probability (figs. hybridizing populations is a plausible driver and E. Zavala for helpful discussions and comments on the
S27 to S32). Given the crucial role of the for the replacements observed in other hy- manuscript. Funding: Q.F. was supported by funding from the
Chinese Academy of Sciences (XDB26000000) and the National
Y chromosome in reproduction and fertility bridization events (27–29).
Natural Science Foundation of China (91731303, 41925009,
and its haploid nature, it is possible that del- 41630102). A.R. was funded by Spanish government (MICINN/
RE FERENCES AND NOTES
eterious mutations or structural variants on FEDER) (grant number CGL2016-75109-P). The reassessment of
1. R. E. Green et al., Science 328, 710–722 (2010). the Spy collection by H.R., I.C., and P.S. was supported by the
the Y chromosome have a larger impact on 2. M. Meyer et al., Science 338, 222–226 (2012). Belgian Science Policy Office (BELSPO 2004-2007, MO/36/0112).
fitness than considered in our simulations. 3. S. Lippold et al., Investig. Genet. 5, 13 (2014). M.V.S., M.B.K., and A.P.D. were supported by the Russian
We therefore refrain from making predictions 4. M. Karmin et al., Genome Res. 25, 459–466 (2015). Foundation for Basic Research (RFBR 17-29-04206). This study
5. I. Olalde et al., Science 363, 1230–1234 (2019). was funded by the Max Planck Society and the European
about the specific process of replacement, be-
6. K. Prüfer et al., Nature 505, 43–49 (2014). Research Council (grant agreement number 694707). Author
cause we lack information about the frequen- 7. C. Posth et al., Nat. Commun. 8, 16046 (2017). contributions: M.P. and J.K. analyzed data. M.H., Q.F., and E.E.
cies of introgressed Y chromosomes in older 8. M. Meyer et al., Nature 505, 403–406 (2014). performed laboratory experiments. H.R., I.C., P.S., L.V.G., V.B.D.,
Neanderthals, potential sex bias in the gene 9. M. Meyer et al., Nature 531, 504–507 (2016). C.L.-F., M.d.l.R., A.R., M.V.S., M.B.K., and A.P.D. provided samples.
10. F. L. Mendez, G. D. Poznik, S. Castellano, C. D. Bustamante, B.V., M.M., and J.K. supervised the project. M.P. and J.K. wrote
flow, and the fitness effects of single-nucleotide Am. J. Hum. Genet. 98, 728–734 (2016). and edited the manuscript with input from all co-authors.
and structural variants on the Y chromosome 11. S. Sawyer et al., Proc. Natl. Acad. Sci. U.S.A. 112, 15696–15700 Competing interests: The authors declare no competing interests.
(26). Nevertheless, our models are a proof-of- (2015). Data and materials availability: Complete source code for
12. K. Douka et al., Nature 565, 640–644 (2019). data processing and simulations, as well as Jupyter notebooks with
principle demonstration that even a simple 13. M. Hajdinjak et al., Nature 555, 652–656 (2018). all analyses and results, can be found at Zenodo (31). Coordinates
difference in the efficacy of purifying selection 14. See supplementary materials. of the capture target regions and sequences of the capture
between two lineages can markedly affect 15. R. E. Wood et al., Archaeometry 55, 148–158 (2013). probes are available at Zenodo (32). All sequence data are available
16. S. Castellano et al., Proc. Natl. Acad. Sci. U.S.A. 111, 6666–6671 from the European Nucleotide Archive under the accession
introgression dynamics of nonrecombining, (2014). number PRJEB39390.
uniparental DNA. 17. Q. Fu et al., Nature 514, 445–449 (2014).
We conclude that the Y chromosomes of 18. S. Mallick et al., Nature 538, 201–206 (2016).
19. K. Prüfer et al., Science 358, 655–658 (2017). SUPPLEMENTARY MATERIALS
late Neandertals represent an extinct lineage 20. M. Kuhlwilm et al., Nature 530, 429–433 (2016). science.sciencemag.org/content/369/6511/1653/suppl/DC1
closely related to modern human Y chromo- 21. M. J. Hubisz, A. L. Williams, A. Siepel, PLOS Genet. 16, Materials and Methods
somes that introgressed into Neanderthals e1008895 (2020). Figs. S1 to S32
22. J. F. Crow, M. Kimura, An Introduction to Population Genetics Tables S1 to S14
between ~370 and ~100 ka ago. The presence
Theory (The Blackburn Press, 1970). References (33–67)
of this Y chromosome lineage in all late Nean- 23. K. Harris, R. Nielsen, Genetics 203, 881–891 (2016). MDAR Reproducibility Checklist
derthals makes it unlikely that genetic changes 24. I. Juric, S. Aeschbacher, G. Coop, PLOS Genet. 12, e1006340
(2016). View/request a protocol for this paper from Bio-protocol.
that accumulated in Neanderthal and modern
25. B. C. Haller, P. W. Messer, Mol. Biol. Evol. 36, 632–637
human Y chromosomes before the introgres- (2019). 9 March 2020; accepted 6 August 2020
sion led to incompatibilities between these 26. S. Colaco, D. Modi, Reprod. Biol. Endocrinol. 16, 14 (2018). 10.1126/science.abb6460
Petr et al., Science 369, 1653–1656 (2020) 25 September 2020 4 of 4
You might also like
- Rectangular Tank Design ROARKS FORMULADocument40 pagesRectangular Tank Design ROARKS FORMULANavasOT100% (3)
- SciAm - 2008 - 299-1!56!63 Traces of A Distant Past (Genetics) StixDocument8 pagesSciAm - 2008 - 299-1!56!63 Traces of A Distant Past (Genetics) StixRenzo Lui Urbisagastegui ApazaNo ratings yet
- Project Sharjah Higher Colleges of Technology: Electrical Power Distribution (ELE-4333)Document5 pagesProject Sharjah Higher Colleges of Technology: Electrical Power Distribution (ELE-4333)Nida NiaziNo ratings yet
- The Complete Mitochondrial DNA Genome of An Unknown Hominin From Southern SiberiaDocument7 pagesThe Complete Mitochondrial DNA Genome of An Unknown Hominin From Southern SiberiaSebastián Ponce ApablazaNo ratings yet
- The Divergence of Neandertal and Modern Human Y ChromosomesDocument8 pagesThe Divergence of Neandertal and Modern Human Y Chromosomeseduviges vegaNo ratings yet
- Nature 09710Document8 pagesNature 09710oldno7brandNo ratings yet
- Gène Neanderthal Chez Non-AfricainsDocument6 pagesGène Neanderthal Chez Non-AfricainsMackenzy DrewNo ratings yet
- Neanderthal and Denisova Genetic AffinitiesDocument12 pagesNeanderthal and Denisova Genetic AffinitiesEmilio Vicente YepesNo ratings yet
- Relethford Origen HombreDocument2 pagesRelethford Origen HombreVictoria SilveiraNo ratings yet
- Ancient DNA and Human History Slatkin 588ed45d8fb66Document8 pagesAncient DNA and Human History Slatkin 588ed45d8fb66Amy MarsNo ratings yet
- Final Draft Anth 790Document11 pagesFinal Draft Anth 790inqu1ryNo ratings yet
- Nuclear DNA From Two Early Neandertals Reveals 80 000 Years of Genetic Continuity in Europe - 5JUN2019 - SCIENCEDocument10 pagesNuclear DNA From Two Early Neandertals Reveals 80 000 Years of Genetic Continuity in Europe - 5JUN2019 - SCIENCEHERAUDNo ratings yet
- Article: Genome-Wide Patterns of Selection in 230 Ancient EurasiansDocument16 pagesArticle: Genome-Wide Patterns of Selection in 230 Ancient EurasiansFeng CaiNo ratings yet
- Piis0092867418301752 PDFDocument19 pagesPiis0092867418301752 PDFlolwut83737No ratings yet
- Piis0092867418301752 PDFDocument19 pagesPiis0092867418301752 PDFlolwut83737No ratings yet
- Prüfer Et Al 2014Document27 pagesPrüfer Et Al 2014Luz Myriam Londoño SalazarNo ratings yet
- Emss 78691Document18 pagesEmss 78691Arthur GonçalvesNo ratings yet
- Genomic Analyses of Pre-European Conquest Human Remains From The Canary Islands Reveal Close Affinity To Modern North AfricansDocument17 pagesGenomic Analyses of Pre-European Conquest Human Remains From The Canary Islands Reveal Close Affinity To Modern North AfricanshasalmorabitNo ratings yet
- Cell, Vol. 90, 19-30, July 11, 1997, Copyright ©1997 byDocument12 pagesCell, Vol. 90, 19-30, July 11, 1997, Copyright ©1997 byjohnny cartinNo ratings yet
- Neanderthal GeneticsDocument7 pagesNeanderthal GeneticsPaul CerneaNo ratings yet
- PIIS0092867400803104Document12 pagesPIIS0092867400803104Adrian Dacosta LazeraNo ratings yet
- 2015 Perry Et Al Insights Into Hominin Phenotypic and Dietary Evolution From Ancient DNA Sequence DataDocument9 pages2015 Perry Et Al Insights Into Hominin Phenotypic and Dietary Evolution From Ancient DNA Sequence DataAlexeyNo ratings yet
- Ancient Human DNA Recovered From A Palaeolithic PendantDocument20 pagesAncient Human DNA Recovered From A Palaeolithic PendantSergio VillicañaNo ratings yet
- Stoneking 1994Document9 pagesStoneking 1994melia243No ratings yet
- Advanced Medicineprize2022Document13 pagesAdvanced Medicineprize2022Venkat TNo ratings yet
- Population Dynamics and Genetic Connectivity in ReDocument39 pagesPopulation Dynamics and Genetic Connectivity in RePutu SulasniNo ratings yet
- Long-Range Organization of Tandem Arrays of A Satellite DNA at The Centromeres of Human ChromosomesDocument5 pagesLong-Range Organization of Tandem Arrays of A Satellite DNA at The Centromeres of Human ChromosomesMariano HorganNo ratings yet
- Volume 40 1 96 6 Neotropical MonkeysDocument11 pagesVolume 40 1 96 6 Neotropical MonkeysAnindyaMustikaNo ratings yet
- Evidencia Centéfica BigfootDocument24 pagesEvidencia Centéfica BigfootMarianCondeNo ratings yet
- More Genomes From Denisova Cave Show Mixing of Early Human GroupsDocument1 pageMore Genomes From Denisova Cave Show Mixing of Early Human GroupsJason Morgan100% (1)
- Reconstructing Human OriginsDocument13 pagesReconstructing Human Originsmilica cucuzNo ratings yet
- PNAS 2014 WildeDocument6 pagesPNAS 2014 Wildepaja.podholkaNo ratings yet
- Primer: Ancient DNA AnalysisDocument26 pagesPrimer: Ancient DNA AnalysisST100% (1)
- Neanderthal-Denisovan Ancestors Interbred With A Distantly Related Hominin - 20FEV2020 - SCIENCEDocument8 pagesNeanderthal-Denisovan Ancestors Interbred With A Distantly Related Hominin - 20FEV2020 - SCIENCEHERAUDNo ratings yet
- Nature 01401Document3 pagesNature 01401Deyvid LuisNo ratings yet
- Genetic Traces of East To West Human ExpDocument9 pagesGenetic Traces of East To West Human Expxavier.quaranteNo ratings yet
- The Eurasian HeartlandDocument6 pagesThe Eurasian HeartlandHakan EnerNo ratings yet
- Jurnal Arthopoda PDFDocument12 pagesJurnal Arthopoda PDFBenny UmbuNo ratings yet
- Unit 2Document23 pagesUnit 2anu ruhan505No ratings yet
- Full Text - Hajdinjak 41586 - 2021 - 3335 - OnlinePDF - S300aDocument20 pagesFull Text - Hajdinjak 41586 - 2021 - 3335 - OnlinePDF - S300aSenneStarckxNo ratings yet
- aYChr-DB A Database of Ancient Human Y HaplogroupsDocument4 pagesaYChr-DB A Database of Ancient Human Y HaplogroupsEranElhaikNo ratings yet
- Díaz Et Al 2016 Genetic Analysis of PaleoDocument23 pagesDíaz Et Al 2016 Genetic Analysis of PaleoJose Vicente Rodriguez CuencaNo ratings yet
- ScholzDocument6 pagesScholzVedad BalesicNo ratings yet
- The Genetic Legacy of The Neanderthals: World Socialist Web SiteDocument2 pagesThe Genetic Legacy of The Neanderthals: World Socialist Web SiteRobert MillerNo ratings yet
- Peerj 04 1556Document36 pagesPeerj 04 1556JanethNo ratings yet
- 1 October 2020Document4 pages1 October 2020RT CrNo ratings yet
- Nihms 931973Document23 pagesNihms 931973sariina.ghNo ratings yet
- Dennis. Human Adaptation and Evolution by Segmental DuplicationDocument9 pagesDennis. Human Adaptation and Evolution by Segmental DuplicationROCIO CERVANTES HERNANDEZNo ratings yet
- American Journal of Physical Anthropology Volume 132 Issue 4 2007 (Doi 10.1002/ajpa.20543) Brian M. Kemp Ripan S. Malhi John McDonough Deborah A. Bolnic - Genetic Analysis of Early Holocene Skel PDFDocument17 pagesAmerican Journal of Physical Anthropology Volume 132 Issue 4 2007 (Doi 10.1002/ajpa.20543) Brian M. Kemp Ripan S. Malhi John McDonough Deborah A. Bolnic - Genetic Analysis of Early Holocene Skel PDFAdrián Mazo-CastroNo ratings yet
- Baca & Molak 2008 DNA Near EastDocument23 pagesBaca & Molak 2008 DNA Near EastvesnadimiNo ratings yet
- Summary and ResultsDocument13 pagesSummary and ResultsDemang GitosantosoNo ratings yet
- Archaeo GeneticsDocument29 pagesArchaeo GeneticsALISTER GARCIA HIMAWANNo ratings yet
- Novel North American Hominins Final PDF DownloadDocument41 pagesNovel North American Hominins Final PDF DownloadRichard Bachman100% (1)
- Lambert & Millar Nature 06Document2 pagesLambert & Millar Nature 06Andrew JonesNo ratings yet
- Week 3.1 The Upper Paleolithic 1 (The Aurignacian)Document25 pagesWeek 3.1 The Upper Paleolithic 1 (The Aurignacian)Ricardo PadillaNo ratings yet
- Near DentalDocument5 pagesNear DentalAna María Villamil CamachoNo ratings yet
- 1 s2.0 S0888754310001552 Main PDFDocument12 pages1 s2.0 S0888754310001552 Main PDFAnonymous sF4FmENo ratings yet
- 3 WiseDocument10 pages3 WiseAnonymous 64x5K7Di9rNo ratings yet
- The Landscape of Neandertal Ancestry in Present-Day Humans: Try Out and Tell Us What You Think.Document20 pagesThe Landscape of Neandertal Ancestry in Present-Day Humans: Try Out and Tell Us What You Think.VargNo ratings yet
- 50 Great Myths of Human Evolution: Understanding Misconceptions about Our OriginsFrom Everand50 Great Myths of Human Evolution: Understanding Misconceptions about Our OriginsNo ratings yet
- The Long Journey of My Little Y Chromosomes: The Origins of One Viking FamilyFrom EverandThe Long Journey of My Little Y Chromosomes: The Origins of One Viking FamilyNo ratings yet
- Analysis On Korea's Soft PowerDocument5 pagesAnalysis On Korea's Soft Powerpeter_yoon_14No ratings yet
- 2539-ArticleText-7142-1-10-201911061Document11 pages2539-ArticleText-7142-1-10-201911061Tamyy VuNo ratings yet
- A Critical Analysis of Cultivation Theory: Original ArticleDocument53 pagesA Critical Analysis of Cultivation Theory: Original ArticleHidya AwalyaNo ratings yet
- Shrum 2017 John Wiley Encycolpedia CultivationDocument13 pagesShrum 2017 John Wiley Encycolpedia CultivationaalijaaproductionsNo ratings yet
- Neanderthals Gene and Modern DiseaseDocument2 pagesNeanderthals Gene and Modern DiseaseTamyy VuNo ratings yet
- Gene Disease EvolutionDocument2 pagesGene Disease EvolutionTamyy VuNo ratings yet
- Parasocial 1Document19 pagesParasocial 1Tamyy VuNo ratings yet
- Physics 101: Fall 2003 Session - General InformationDocument4 pagesPhysics 101: Fall 2003 Session - General InformationMAnohar KumarNo ratings yet
- HW4 SolDocument5 pagesHW4 SolLidia Monica AnwarNo ratings yet
- LSM SSD Firmware Update 020811-R00Document3 pagesLSM SSD Firmware Update 020811-R00Nyser ZegnaNo ratings yet
- V-Wheels (25203-02)Document33 pagesV-Wheels (25203-02)nicopoehlmannNo ratings yet
- Hyperreal: A Hypermedia Model For Mixed Reality: Luis Romero, Nuno CorreiaDocument9 pagesHyperreal: A Hypermedia Model For Mixed Reality: Luis Romero, Nuno CorreiaVideaux LondonNo ratings yet
- Topic 5 (Updated) Gravimetric Methods of AnalysisDocument38 pagesTopic 5 (Updated) Gravimetric Methods of AnalysisAdznaira AmilussinNo ratings yet
- Water Cooling TowerDocument1 pageWater Cooling Toweryadi_baeNo ratings yet
- mv110 120 130Document2 pagesmv110 120 130Jorge Martín Cabrera RochaNo ratings yet
- Termostato HoneywellDocument48 pagesTermostato HoneywellTámesis LA Tierra Del Siempre VolverNo ratings yet
- Creative Media 17-06 PDFDocument21 pagesCreative Media 17-06 PDFsinduja.cseNo ratings yet
- L7CN :connecting LANs, Backbone Networks, and Virtual LANsDocument12 pagesL7CN :connecting LANs, Backbone Networks, and Virtual LANsAhmad Shdifat100% (1)
- Fundamentals of Arcgis Desktop PathwayDocument28 pagesFundamentals of Arcgis Desktop PathwayhujabryNo ratings yet
- 16 - NeuronDocument2 pages16 - NeuronSimonNo ratings yet
- Sample Question Paper Class - X Session - 2021-22 Term 1 Subject-Mathematics (Standard) 041Document7 pagesSample Question Paper Class - X Session - 2021-22 Term 1 Subject-Mathematics (Standard) 041hweta173No ratings yet
- RM Practical FileDocument59 pagesRM Practical Filegarvit sharmaNo ratings yet
- Example of Ferrule Selection ChartDocument1 pageExample of Ferrule Selection ChartKyi HanNo ratings yet
- Rough Neutrosophic Relation On Two Universal SetsDocument14 pagesRough Neutrosophic Relation On Two Universal SetsMia AmaliaNo ratings yet
- Syl 2372Document8 pagesSyl 2372Ismael 8877No ratings yet
- Hipass PDFDocument1 pageHipass PDFahmed_galal_waly1056No ratings yet
- Blast Mitigation Using Water-A Status Report: Naval Research LaboratoryDocument50 pagesBlast Mitigation Using Water-A Status Report: Naval Research Laboratoryigor VladimirovichNo ratings yet
- Intercepts, Zeroes, and AsymptotesDocument12 pagesIntercepts, Zeroes, and AsymptotesMoses Zackary TolentinoNo ratings yet
- Deep Sea Electronics: Model 5220 Installation and Configuration InstructionsDocument2 pagesDeep Sea Electronics: Model 5220 Installation and Configuration Instructionsdhani_is100% (1)
- Q2 Tech - Draw8 M4Document38 pagesQ2 Tech - Draw8 M4Savannah VinluanNo ratings yet
- Rotor-Dynamics Analysis ProcessDocument16 pagesRotor-Dynamics Analysis ProcessLaksh ManNo ratings yet
- Iso 2896 - 2001 eDocument16 pagesIso 2896 - 2001 ehappyhandslearningcenter2023No ratings yet
- 100 Practical Exercises in Music Composition 0190057246 9780190057244 Compress 3Document45 pages100 Practical Exercises in Music Composition 0190057246 9780190057244 Compress 3Lê Thanh XuânNo ratings yet
- Book List For ParamaDocument6 pagesBook List For ParamaShafayet UddinNo ratings yet
- Estimation of Cu & ZNDocument7 pagesEstimation of Cu & ZNjhfgh67% (3)