Professional Documents
Culture Documents
Eukaryotic Gene Transcription Notes
Eukaryotic Gene Transcription Notes
Uploaded by
EshaOriginal Description:
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Eukaryotic Gene Transcription Notes
Eukaryotic Gene Transcription Notes
Uploaded by
EshaCopyright:
Available Formats
lOMoARcPSD|8427367
Chapter 4 PDF process of transcription
Eukaryotic gene control and development 351 (University of Pretoria)
StuDocu is not sponsored or endorsed by any college or university
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Introduction
§ Gene regulation primarily operates at the level of transcription –
determines which genes will be transcribed into RNA in specific tissues
or in response to specific stimuli.
Ø Operates at the level of chromatin structure so that the DNA that is to
be transcribed moves to a more open chromatin structure allowing
access to regulatory molecules.
Ø RNA polymerases can copy the DNA into RNA together with a variety of
transcription factors that can stimulate o inhibit polymerase activity.
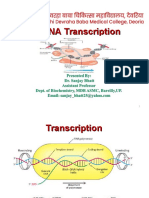
Ø Eukaryotic transcription = synthesis of RNA molecules copied from
template DNA. Carried out in the nucleus.
Different types of RNA are transcribed in cells:
The 3 stages of transcription:
§ STAGE 1: Initiation
Ø Requires open chromatin at promoter to allow access to regulatory
molecules
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø Transcription factors (TFs) locate and bind to short DNA control
regions in promoter regions.
Ø General TFs recruit RNA pol to promoter
Ø Additional activator/repressor proteins determine level of
transcription
§ STAGE 2: Elongation
§ STAGE 3: Termination
4.1) TRANSCRIPTION BY RNA POLYMERASES
§ RNA polymerases: Enzymes that is capable of using the DNA as a
template so that a complementary RNA copy is produced by the
polymerization of ribonucleotides.
Ø In Prokaryotes= Single RNA polymerase
Ø In Eukaryotes= 3 RNA polymerases found in the nucleus (RNA pol I, II
and III)
Ø In plants = 2 RNA polymerases (RNA IV and V) which produce
inhibitory chromatin structure via siRNAs.
Ø RNA pol I, II and III are all large multi-subunit enzymes, however, they
can be distinguished via sensitivity to the fungal toxin α-amanitin and
their activity on distinct sets of genes.
Ø RNA pol II= in protein-encoding genes and genes for siRNAs
Ø RNA pol I= Genes encoding rRNAs (28S, 18S and 5.8S)
Ø RNA pol III= Genes encoding tRNAs and 5S rRNA
Transcription initiation by RNA polymerase II
§ Different genes transcribed by RNA pol II have an AT rich sequence 30
nucleotides upstream from the start site (located at +1)
§ TATA box – plays a role in promoting initiation of transcription and in
positioning the start site of transcription for RNA pol II.
Ø Deletion= no transcription
Ø Initial DNA target site for the assembly of the basal pre-initiation
transcription complex for RNA pol II.
Ø Transcription factors bind to the complex in a defined order along with
the RNA pol to form the basal transcription complex (pre-initiation
complex or PIC)
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
§ Formation of the Basal complex:
Ø STEP A: TFIID binds to the TATA via its TBP
subunit. - Facilitated by the presence of TFIIA
Ø STEP B: TFIID/DNA complex is recognized by
TFIIB, which binds, on the opposite side of the DNA
to which TFIIA is bound. TFIIB contacts special
sequences on either side of the TATA box called
upstream and downstream BRE’s.
Ø STEP C: TFIIB recruits RNA pol II that is in
association with TFIIF. TFIIB helps to position RNA
pol II.
Ø STEP D: TFIIE and TFIIH associate to form the full
PIC. TFIIH has kinase activity capable of
phosphorylating the C-terminal domain of RNA pol
II’s largest subunit, which is known as RPBI.
Phosphorylation allows initiation to occur, it plays a
role in both transcription and DNA repair
Ø STEP E: RNA pol II and TFIIF move along strand
to produce transcript. TFIIA and TFIID remain
behind for repeated rounds of transcription.
TFIID is a large multi-component protein complex
TFIID associates
with other TFs
such as TFIIB
and TFIIA
§ TFIID is composed of many different TFs
§ TATA-binding protein (TBP) – Binds to the TATA box, thus TBP is a key
component of the initial DNA-binding complex.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø TBP-associated factors (TAF’S) – Allows TFIID to respond to activators
§ Molecular clamp structure: domains form a groove that fits DNA
§ DNA is transcribed into RNA in the interior of
the Polymerase molecule; it encounters a
wall of protein within the RNA pol. This
forces it to make a right-angled turn,
exposing the end of the nascent RNA and
allowing ribonucleoside triphosphates to be
added to it as transcription occurs.
§ The DNA-RNA hybrid encounters another
part of the RNA pol known as the rudder,
which forces the separation of the RNA from
the DNA. This allows the newly formed RNA
to exit and the dsDNA to reform.
§ RNA pol Holoenzyme: Some RNA pol found within the cell is
associated with large
Number of proteins
including TFIIB, TFIIF and
TFIIH.
Ø Components in the
Holoenzyme are involved in
opening up the chromatin
structure and allowing the
polymerase complex to be
stimulated by transcriptional
activators.
Ø An alternative pathway
exists where TFIIA and
TFIID can recruit the
holoenzyme complex and
RNA Pol.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Deletion mutation analysis and DNase1 footprinting
§ The entire upstream region of a gene could be deleted with no effect on
gene expression.
Ø Deletions allow us to identify control regions
Ø In the 5S rRNA gene – deletion had no effect until a boundary 40 bases
within the transcribed region was crossed. The internal control region
of the transcription of the 5S rRNA gene was identified within the
transcribed region.
§ Using particular transcription factors (in the 5S rRNA case TFIIIA
was used) we can identify control regions.
Saddle-like structure of TBP bound to TATA box
§ TBP binding bends the DNA so that it follows the curve of a saddle
structure within TBP.
§ The structure of TFIID bound to DNA resembles the core nucleosome.
Ø TFIID bends the DNA at the promoter in a similar way to the bending
of DNA around the nucleosomes in the remaining DNA
Ø The TAFII250 subunit of TFIID has histone acetyltransferase activity
Allowing it to modulate chromatin structure by acetylating
histones.
4.2)TRANSCRIPTIONAL ELONGATION AND TERMINATION
Transcriptional Elongation requires further phosphorylation of RNA
polymerase II
§ Phosphorylation of serine 5 within the C-terminal repeat region in the
largest subunit of RNA pol II results in transcription of the gene.
§ The first nucleotide in the DNA template that is copied into RNA (the +1
nucleotide) is usually a C or T residue (therefore the RNA starts with a
G or an A)
§ The nucleotides in the chain are joined via phosphodiester bonds
§ The chain grows in a 5’ to 3’ direction
§ Following transcriptional initiation, transcription proceeds for about
20-30 bps and then polymerase pauses.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø Release of this block and continued transcriptional
elongation requires phosphorylation of the C-terminal
repeat region of the RPB1 subunit of RNA pol II on
Serine 2 (S2) of the Tyr-Ser-Pro-Thr-Ser-Pro-Ser
repeated sequence.
Ø The phosphorylation of S2 is linked to the modification
of the free 5’ end of the nascent RNA transcript by
addition of a modified G nucleotide in a process known
as capping
Ø Capping occurs when the RNA transcript is 20-30
bps long and promotes the binding of the pTEF-b
kinase protein.
Ø The pTEF-b kinase then phosphorylates the C-
terminal repeat of RNA pol II on S2 allowing
elongation to occur.
Ø Polymerase pausing is widespread and represents a
potential control point for the regulation of
transcription
Termination of transcription occurs downstream of the
polyadenylation signal
§ The 3’ end of the mature mRNA is defined by the post-transcriptional
addition of a run of adenosine nucleotides to produce a Poly (A) tail in
the process known as polyadenylation.
Ø This process involves the recognition of a polyadenylation signal
including the invariant sequence AAUAAA within the RNA followed by
internal cleavage of the RNA downstream of this sequence and
polyadenylation of the free 3’ end.
§ Mutation of the polyadenylation signal leads to interference with
normal termination.
§ Polyadenylation facilitates termination in several ways:
Ø The cleavage / polyadenylation protein complex which binds to the
transcribing RNA polymerase recruits Rat 1 exonuclease enzymes.
Ø Polyadenylation complex produces the mature mRNA and a
downstream RNA with a free 5’ end. The Rat 1 binds to the free 5’ end
and degrades it.
Ø The binding of the polyadenylation complex slows down the rate of
transcription by the polymerase and make it pause frequently so Rat 1
can catch up
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø Therefore, the recruitment of Rat 1, the slowing of transcription and
the cleavage at the RNA transcript during polyadenylation that
generates a free 5’ end RNA all plays a role in termination.
4.3) THE GENE PROMOTER
§ A number if genes transcribed by RNA pol II contain a TATA box 30
bps upstream of the transcriptional start site.
§ Other RNA pol-II transcribed genes utilize an initiator (Inr) sequence
with the consensus 5’-YCANTYY-3’ with the A being the first nucleotide
transcribed (Y is C or T and N is any nucleotide) this sequence is
located around the transcriptional start site.
§ TBP is recruited to these promoters via binding of TAFs.
§ Most genes have a TATA box, an Inr sequence and TFIIB-binding sites
(BREs) both upstream and downstream.
§ A small number of genes lack both a TATA box and an Inr sequence,
but they have a CpG island close to the transcriptional start site and
have a low level of transcription with a variable transcription start site.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
§ Core/Basal Promoter- Region around or just upstream of the
transcription start site containing the TATA box and/or Inr sequence as
well as the BREs. Serves to recruit the basal transcription complex.
Ø The core promoter is the minimal region that can direct initiation of
transcription.
Ø Even in the presence of a TATA box or an Inr sequence the level of
transcription will be relatively low –Increased by the presence of
upstream promoter elements, which can enhance the
stability, or activity of the basal complex.
§ Promoter= core promoter and upstream promoter elements.
§ The production of a protein-coding RNA transcript is generally
unidirectional, but in some cases a noncoding RNA is produced in the
opposite direction and thus the transcription is bidirectional.
The 70kDa heat-shock protein gene contains a typical promoter for RNA
polymerase II
§ When a wide variety of cells are exposed to an increase in temperature
there is an increased synthesis of heat shock proteins- Hsp70 is the
most abundant.
Ø Increased synthesis is mediated by increased transcription of the
corresponding gene.
§ The gene has several promoter elements including TATA, CCAAT, Sp1
(GC box), CRE (cAMP response element) and AP2 (Activator protein 2)
boxes
Ø Each element is recognized by a specific TF of the basal complex to
initiate transcription.
Ø Two copies of the Sp1 box are found in the Hsp70 gene promoter, This
CG-rich DNA sequence binds a transcription factor known as Sp1,
which is present in all cell types.
Ø The CCAAT box is located upstream of the start site of a wide variety of
genes – plays an important role in allowing transcription by binding
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Constitutively expressed factors such as CTF (CAAT transcription factor)
The heat shock element is found only in heat-inducible genes
§ Found in Hsp70 gene promoter.
§ Found 62 bp upstream of the transcription start site in the Drosophilia
Hsp70 gene.
§ Plays a critical role in mediating the observed heat inducibility of
transcription of these genes.
§ HSE is recognized by heat-shock factor protein (HSF)
§ HSE does not possess an inherent temperature sensor
§ HSE allows these genes to respond to elevated temperatures via
increased transcription of these genes.
§ Tested via a Reporter gene- encodes a protein whose amount can be
readily quantified (eg β-galactosidase, luciferase etc.)
Ø The regulatory sequence, which is being tested, is linked to the
reporter gene and introduced into cells so that the effect of the
regulatory sequence on production of the protein encoded by the
reporter gene can be measured.
Ø Can be used to link an entire promoter to a reporter gene, the effect of
a short DNA sequence on the activity of a control promoter linked to a
reporter gene (HSE) and the effect of more distant regulatory
sequences such as enhancers on a control promoter.
§ In un-induced cells: TBP is bound to the TATA box and TF known as
the GAGA factor is bound to an upstream GA rich site. The binding of
GAGA displaces a nucleosome and creates DNase1 hypersensitive sites
– Open chromatin structure so the gene has potential to be activated.
§ In induced cells: HSF binds to HSE to activate transcription. HSF is pre-
formed but inactive in cells, Heat induction changes its shape and
allows binding to HSE.
Other response elements are found in the promoters of genes with
different patterns of expression
§ These sequences bind specific proteins, which are
synthesized/activated in response to the inducing signal.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
§ Many of the sequences exhibit dyad symmetry which is a similar
sequence being found in the 5’ to 3’ direction on each strand –
Therefore they bind to the site in dimeric form (consists of 2 protein
molecules)
§ One gene can have multiple patterns of regulation – allows induction of
a particular gene in response to a given stimulus.
The proteins binding to short DNA sequence elements can be
characterized by a variety of techniques
a) The DNA mobility shift assay
§ The DNA sequence (which is radioactively labeled) and cell extract
are mixed. The DNA moving more slowly in gel electrophoresis
detects protein binding to DNA.
§ Protein binding to DNA results in the appearance of a retarded
band, which will be absent if no protein in the test extract, can bind.
§ Also known as band shift or gel retardation assay
§ Method:
Ø STEP 1: Radioactively label DNA containing the DNA sequence of
interest.
Ø STEP 2: Mix with the whole cell or nuclear extract and incubate
Ø STEP 3: Run on the mixture on a nondenaturing
electrophoresis gel and observe the position of the radioactive
bands by autoradiography
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø STEP 4: Detect protein banding of DNA by appearance of the
retarded band in the autoradiograph
§ Can be used to see if the presence or absence of the DNA binding-
activity correlates with the pattern of gene activity conferred by the
DNA sequence.
§ Possible to characterize DNA-binding specificity of the binding
factor by adding an unlabeled competitor with the labeled DNA of
interest – If the unlabeled competitor can also bind then there will
be no band.
§ “Super-shifted” complex –antibody added to the assay to see if the
DNA-binding factor binds, the complex has lower mobility.
b) DNase1 footprinting assay
§ A binding of a protein protects its binding site in DNA from
digestion.
§ Used to identify where protein binds within the DNA sequence and
its position relative to other proteins binding to adjacent DNA
sequences.
§ Method:
Ø STEP 1: Radioactively label DNA fragment containing the
sequence of interest at one end only.
Ø STEP 2: Mix with whole cell or nuclear extract and incubate.
Ø STEP 3: Briefly digest mixture with DNase1 to produce a series of
DNA fragments each differing in length by a single nucleotide.
Ø STEP 4: Run the DNA fragments on denaturing polyacrylamide
gel capable of resolving fragment lengths and detecting binds by
autoradiography.
Ø STEP 5: Visualize area where protein was bound at gap in the
ladder of the DNA fragment – protects region of DNA from digestion.
§ Delineates nucleotides actually bound by binding protein.
§ Possible to visualize multiple footprints on a single DNA molecule,
thereby elucidating the pattern of proteins bound to the promoter or
regulatory region of a particular gene.
§ Valuable in examining the interaction of proteins with particular
DNA sequences.
c) Chromatin Immunoprecipitation
§ Examines TF binding in intact cells
§ Antibodies to a particular TF is used to immunoprecipitate and purify
DNA fragments to which it is bound within normal chromatin structure.
§ Method:
Ø STEP 1: Fix living cells with formaldehyde to stably cross-link
transcription factors to DNA-binding sites
Ø STEP 2: Fragment chromatin into small fragments and purify
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø STEP 3: Immunoprecipitate the TF of interest and its target DNA,
using the antibody to the TF
Ø STEP 4: Break DNA-protein cross-links and isolate DNA
Ø STEP 5: Characterize isolated DNA
§ Used to detect if a gene of interest is present in the immunoprecipitate
under certain conditions.
§ Can be used with DNA arrays or gene chips to array DNA of a particular
genome on a glass slide = ChIP-chip analysis
Promoter regulatory elements act by binding factors which either affect
chromatin structure and/or influence transcription directly
§ Short DNA sequence elements located near the transcription start site
play an important role in controlling
Mediating transcriptional activation
by binding a specific protein does
this Gene expression-.
Ø (a) Binding of a specific protein may
result in displacement of a
nucleosome and a generation of a
DNase1 hypersensitive site, allowing
the TFs to access the gene easily (eg
Glucocorticoid receptor)
Ø (b) The direct activation of
transcription by DNA binding
proteins- Protein binds and interacts
with other proteins that are already
bound at the promoter, this causes
the formation of stable complex
leads to an increase in transcription
(eg HSF binds to TBP of RNA pol)
§ These 2 mechanisms are not
exclusive:
Ø Glucocorticoid receptors also contain an activation domain capable of
interacting with other DNA-bound TFs.
Ø HSF can induce acetylation of histone H4 which results in a more open
chromatin structure-also stimulates basal transcriptional complex
4.4) ENHANCERS AND SILENCERS
Enhancers are regulatory sequences that act at a distance to increase
gene expression
§ Lack promoter activity and are unable to direct transcription
themselves
§ Increases transcription dramatically by enhancing promoter activity
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
a. An enhancer element can activate a
promoter when placed up to several
thousand bases from the promoter
b. An enhancer can activate a promoter
when placed in either orientation
relative to the promoter
c. An enhancer can activate a promoter
when placed upstream or downstream
of the transcribed, or within an
intervening sequence, which is
removed from the RNA by splicing.
Many enhancers have cell-type or tissue-
specific activity
§ Active only in the tissue in which the
gene from which they were derived- not
in other tissues.
§ Plays a key role in mediating the
observed pattern of gene regulation.
§ Clone enhancer is upstream of a
reporter gene where the reporter gene
Is under control of an unrelated promoter.
§ Transfect into different cell types.
§ Tissue-specific enhancer will activate its own promoter and the
promoter of the reporter gene in tissue type A but not in other tissues.
§ Eg) Tissue enhancer specificity in genes encoding antibodies, specific
cancers and genes specifically expressed in the liver.
§ Tissue-specific activity of these enhancer elements is likely to play a
crucial role in observed tissue-specific pattern of excretion of the
corresponding genes.
SV40 virus gene linked to enhancer of insulin
§ Insulin shows tissue-specific expression in pancreas, expression is
under control of a pancreas-specific enhancer.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
§ Experiment: replaced insulin coding region in the mouse genome with
virus gene X – made a transgenic mouse. Hybrid genes thus presents in
all mouse cells.
§ When analyzed, virus gene X is only expressed in the pancreas=
pancreas-specific promote and enhancer still present upstream of
hybrid gene in all mouse cells.
§ In many cases, the tissue-specific expression of a gene will be
determined both by the enhancer element and sequences adjacent to
the promoter.
Enhancers influence transcription both by recruiting DNA-binding
transcription factors and by altering chromosome structure
§ Enhancers show sequence similarity to elements of the promoter.
§ Often have multiple sequence motifs similar to promoters, to
which regulatory protein factors can bind.
§ These repeat sequences function together for enhanced activation.
§ The array of different proteins that assemble on the enhancer is called
an enhanceosome
§ Role – enhancers increase transcription by increasing the concentration
of TF’s (activators) in the vicinity of the promoter.
§ Similar mechanisms as promoter elements:
Ø By changing chromatin structure, leading to nucleosome displacement
and the formation of hypersensitive sites.
Ø By direct interaction with proteins of the basal transcription apparatus
factors
§ Enhanceosome:
Ø A complex of the enhancer DNA element with activators contacting this
DNA
Ø Helps to bend DNA so that it may interact with other proteins
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
A model for enhancer action
§ Enhanceosome complex binds to an enhance-consists
of 8 different proteins including DNA-binding protein
HMGI (Y), activators and RNA pol II.
§ The enhanceosome complex first recruits a histone
acetyltransferase complex stimulating the recruitment
of chromatin remodeling complex SWI/SNF.
§ The chromatin is opened up to allow the binding of
RNA polymerase II and associated proteins.
§ Nucleosome is displaced and binding of basal
transcriptional complex.
The “at a distance” action of enhancers involves both
DNA-binding and transcription factors and alteration of
chromatin structure
§ In the case of chromatin structure: changes caused by a
Protein binding to enhancer could be propagated over large distances
in both directions, resulting in the observed distance, position and
orientation independence of the enhancer.
§ DNase1 hypersensitivity sites are present in
enhancer elements.
§ The nucleosome-free gap in the DNA of SV40 is in the
enhancer.
§ Distant enhancers can activate promoters by looping
of the intervening DNA.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
§ This allows protein bound to an enhancer to interact
with proteins bound at the basal promoter.
§ Proteins bound to the enhance can also bend DNA to
allow interaction at basal promoter.
§ In some instances, DNA-binding activators bound to
enhancer can recruit the basal transcription complex
(TFIID, TFIIB, RNA pol II) to the enhancer itself. In
response to an activation signal. The basal complex is
then transferred to promoter to initiate transcription.
Ø Poises the gene for transcription
Ø When the cell receives an activation signal, the basal
complex is then transferred to the promoter to initiate
transcription.
Ø Recruitment of the basal transcriptional complex via an
enhancer may therefore occur particularly in genes that
lack promoter elements that would normally recruit the
complex.
Many enhancers are transcribed to long ncRNA’s
§ The transcriptions patterns of these RNAs occur in
situations where the enhancer is activated and the
regulatory RNA stimulates enhance activity.
§ (a) Could result in the recruitment of factors which
result in more open chromatin structure of the
enhancer, this allows the binding of regulatory proteins
which stimulate promoter activity.
§ (b) Chromatin structure at the promoter can also be
affected. This likely involves the looping of DNA – leads
to a long noncoding RNA that is transcribed from the
enhancer coming into close contact with the promoter
and recruiting proteins that can alter the chromatin
structure and enable transcription of the gene.
Silencers can act at a distance to inhibit gene expression
§ Some enhancer sequences act in a negative manner,
inhibiting the expression of genes, which contain
them.
§ First identified in cellular oncogene (c-myc)
§ Silencers- genes encoding proteins as diverse as
collagen type II, growth hormone and glutathione
transferase P.
§ Can act on distant promoters when present in either
orientation.
§ Silencers can:
Ø (a) Recruit proteins that direct a closed chromatin
Conformation in the region of the gene. Proteins bind to the silencer.
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Ø (b) Recruit proteins that directly inhibit transcription by interacting
with RNA polymerase and associated Transcription factors. The TF has
a negative effect on promoter activity.
§ Inhibition by the Polycomb Transcription factors:
Ø The binding of Polycomb to a silencer element recruits a histone
methyl-transferase enzyme.
Ø Induces Ubiquitination of H2A and the methylation of H3 on lysines
9 and 27- this promotes tightly packed chromatin.
Ø Inactivation of Polycomb by mutation results in aberrant activation of
specific genes.
PRC-Polycomb TF’s (repressors)
PRE- Polycomb response elements (silencer) Methylation
Each gene contains multiple response elements:
§ The regulatory region of a human metallothionein gene
contains regulatory elements in the promoter and enhancer.
Ø Promoter: elements for metal induction.
Ø Enhancer: element for response to glucocorticoid
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
lOMoARcPSD|8427367
Recombinant DNA techniques have been used to alter the orientation
and location of DNA control elements to study the effect of the change on
transcription levels
a. Core promoter alone= basal
level of transcription
b. Removal of core promoter= no
Transcription
c. Enhancer alone= cannot
substitute activity of core
promoter
d. Enhancer+ core promoter=
Higher levels of transcription
e. Enhancer moved upstream=
Increased transcription
f. Inverted orientation of
enhancer= increased
transcription
g. Enhancer moved to 3’ end
of structural gene=
increased transcription
Downloaded by Esha Galaiya (e.galaiya3322@gmail.com)
You might also like
- BIOL 200 Molecular Biology Lecture NotesDocument44 pagesBIOL 200 Molecular Biology Lecture NotesDantong JiaNo ratings yet
- Gamsat Sample Test - PregenieDocument31 pagesGamsat Sample Test - PregenieBilly Robins100% (1)
- Chapter 1 Iq RQ AnswersDocument14 pagesChapter 1 Iq RQ AnswersFDSNo ratings yet
- Transcription in EukaryotesDocument49 pagesTranscription in EukaryotesmrlabdesigneNo ratings yet
- Regulation of Gene Expression in Eukaryotes: Presented by Quratulain (19-Arid-1403) Asadullah (19-Arid-1379)Document32 pagesRegulation of Gene Expression in Eukaryotes: Presented by Quratulain (19-Arid-1403) Asadullah (19-Arid-1379)Beauty LiciousNo ratings yet
- RNA Transcription and TranslationDocument11 pagesRNA Transcription and TranslationMaiSakurajimaNo ratings yet
- Surgery of Transcription InsightDocument19 pagesSurgery of Transcription Insightnhung_ducuments8261No ratings yet
- L1 TranscriptionDocument13 pagesL1 TranscriptionvarasamaliaNo ratings yet
- RNA Metabolism: Durriya Naeem KhanDocument26 pagesRNA Metabolism: Durriya Naeem KhanMniaz KhanNo ratings yet
- Transcription in Prokaryotes: Single RNA PolymeraseDocument29 pagesTranscription in Prokaryotes: Single RNA PolymeraseShubhamNo ratings yet
- RNA ReplicationDocument23 pagesRNA ReplicationDharaneeshwari Siva-F&NNo ratings yet
- Science FairDocument41 pagesScience FairSafanaNo ratings yet
- Transcription and Regulation of Gene Expression: By: Lyka Marie C. Falcasantos BSN - 1DDocument16 pagesTranscription and Regulation of Gene Expression: By: Lyka Marie C. Falcasantos BSN - 1DRemzAbdullaNo ratings yet
- Dna L12 NotesDocument6 pagesDna L12 NotesellieNo ratings yet
- TranscriptionDocument100 pagesTranscriptionSreshttNo ratings yet
- UNIT 7 Eukaryotic TranscriptionDocument10 pagesUNIT 7 Eukaryotic TranscriptionSarah PavuNo ratings yet
- Chapter 5 Protein SynthesisDocument26 pagesChapter 5 Protein SynthesisT MokshithaNo ratings yet
- Euk TranscriptionDocument22 pagesEuk TranscriptionManish DasNo ratings yet
- TranscriptionDocument64 pagesTranscriptionmadhura480No ratings yet
- Transcripcion EucariotaDocument21 pagesTranscripcion EucariotaAntoniaNo ratings yet
- RNA Synthesis and SplicingDocument64 pagesRNA Synthesis and SplicingLaurine PigossoNo ratings yet
- Gene ExpressionDocument42 pagesGene ExpressionggsbwctfgbNo ratings yet
- Presentation Print TempDocument37 pagesPresentation Print TempDreamcatcher DreamcatcherNo ratings yet
- DNA TranscriptionDocument25 pagesDNA TranscriptionAkss ShauryaNo ratings yet
- Biochemistry: RNA Synthesis and ProcessingDocument54 pagesBiochemistry: RNA Synthesis and ProcessingAqsa YaminNo ratings yet
- Lecture8 DNA-Dependent RNA Synthesis-2Document50 pagesLecture8 DNA-Dependent RNA Synthesis-2Constance WongNo ratings yet
- TranscriptionDocument34 pagesTranscriptiondrhydrogenNo ratings yet
- 11 Chapter 5Document38 pages11 Chapter 5toobashafiNo ratings yet
- Draft 1Document11 pagesDraft 1Afiah LutfiNo ratings yet
- 5 11transcription-2013Document30 pages5 11transcription-2013jernsssNo ratings yet
- Transcription ProkaryoticDocument30 pagesTranscription ProkaryoticDibya Jyoti ParidaNo ratings yet
- Bala Sir Transcrption PDFDocument9 pagesBala Sir Transcrption PDFAnjali Ak GuptaNo ratings yet
- Transcription in ProkaryotesDocument7 pagesTranscription in Prokaryotesmeprabhatmishra99No ratings yet
- TranscriptionDocument10 pagesTranscriptionHardik ManekNo ratings yet
- Chapter 7 Part 1Document26 pagesChapter 7 Part 1coolsuernameNo ratings yet
- SYBT TranscriptionDocument69 pagesSYBT TranscriptionMeir SabooNo ratings yet
- Rna Biosynthesis (Transicription)Document33 pagesRna Biosynthesis (Transicription)Alaa AlmajedNo ratings yet
- TranscriptionDocument61 pagesTranscriptiondeepak mauryaNo ratings yet
- RNA and Transcription (Gene Expression) : The Central DogmaDocument21 pagesRNA and Transcription (Gene Expression) : The Central DogmaAnonymous tgeYTDuzTbNo ratings yet
- Transcription: - by - S. Sivaranjani Arunnehru - Assistant Professor - Bon Secours College For Women - ThanjavurDocument33 pagesTranscription: - by - S. Sivaranjani Arunnehru - Assistant Professor - Bon Secours College For Women - ThanjavurGayathri deviNo ratings yet
- Gene RegulationDocument16 pagesGene RegulationMarkrobert MagsinoNo ratings yet
- Transcriprion ManuscriptDocument14 pagesTranscriprion ManuscriptAsif AhmedNo ratings yet
- MB Chapter 5.2. Eukaryotic TranscriptionDocument39 pagesMB Chapter 5.2. Eukaryotic TranscriptionMustee TeferaNo ratings yet
- Messenger Rnas (Mrnas) : This Class of Rnas Are The Genetic Coding TemplatesDocument8 pagesMessenger Rnas (Mrnas) : This Class of Rnas Are The Genetic Coding TemplatesRavi AlugubelliNo ratings yet
- Physio Protein SynthesisDocument3 pagesPhysio Protein SynthesisM.TAYYAB KHANNo ratings yet
- BMM GM2 ADocument6 pagesBMM GM2 ASleepyHead ˋωˊNo ratings yet
- Report in Cell BiologyIIDocument3 pagesReport in Cell BiologyIIGenessa Agustin BuenafeNo ratings yet
- Chemistry Lec.22 (Nucleic Acids 6)Document6 pagesChemistry Lec.22 (Nucleic Acids 6)Muhammed AbdulsamadNo ratings yet
- Transcription and RNA Processing in EukaryotesDocument13 pagesTranscription and RNA Processing in EukaryotesAbdulfattah NoorNo ratings yet
- Kuliah 8. Transcription (Oke)Document43 pagesKuliah 8. Transcription (Oke)Sinta DosiNo ratings yet
- Eukaryotic Transcription ProcessDocument1 pageEukaryotic Transcription ProcessSomNo ratings yet
- Transcription and TranslationDocument2 pagesTranscription and TranslationRobert Martin PuertaNo ratings yet
- Transcription and TranslationDocument2 pagesTranscription and TranslationRobert Martin PuertaNo ratings yet
- Transcription NotesDocument2 pagesTranscription Notesmartinc08No ratings yet
- Lec 7 TranscriptionDocument30 pagesLec 7 TranscriptionYasmin BalochNo ratings yet
- General Transcription FactorsDocument12 pagesGeneral Transcription Factorssonuqazx6900No ratings yet
- Transcription in Prokaryotes and Eukaryotes: (Frequently Asked Questions)Document6 pagesTranscription in Prokaryotes and Eukaryotes: (Frequently Asked Questions)Asif AhmedNo ratings yet
- 21 TranscriptionDocument39 pages21 TranscriptioneraasyahirahNo ratings yet
- Topic 10 Transcription Lecture NotesDocument33 pagesTopic 10 Transcription Lecture NoteskambulukatambilaiNo ratings yet
- Decker 1Document9 pagesDecker 1Katarina Kaca GacevicNo ratings yet
- Molecular Biology: Unit 3Document64 pagesMolecular Biology: Unit 3John Rajan ThavamNo ratings yet
- RNA Synthesis, Processing SBT 211Document59 pagesRNA Synthesis, Processing SBT 211TADIWANASHE TINONETSANANo ratings yet
- Biomolecule Biomolecule Biomolecule Biomolecule Biomolecules SS SSDocument15 pagesBiomolecule Biomolecule Biomolecule Biomolecule Biomolecules SS SSOm AgrawalNo ratings yet
- ZOO 114 DNA Function LectureDocument53 pagesZOO 114 DNA Function Lecturerakingbulugbe194No ratings yet
- BMB7010 Exam Answers Dec07Document5 pagesBMB7010 Exam Answers Dec07nomansnNo ratings yet
- 04 Macromolecules-1Document53 pages04 Macromolecules-1keikisboyNo ratings yet
- 11th STD BIO-BOTANY Revised English Medium Book Back AnsDocument51 pages11th STD BIO-BOTANY Revised English Medium Book Back AnsDeepsha online100% (1)
- Emboj00111 0098Document8 pagesEmboj00111 0098haddig8No ratings yet
- Study Guide For Exam 1 F20Document4 pagesStudy Guide For Exam 1 F20Adam KatzNo ratings yet
- 2.7 DNA Replication, Transcription, and TranslationDocument25 pages2.7 DNA Replication, Transcription, and TranslationJennNo ratings yet
- H1 Revision Notes DNA and GenomicsDocument6 pagesH1 Revision Notes DNA and GenomicsJiaLi XieNo ratings yet
- Chem41 Lab Manual S15 v3Document53 pagesChem41 Lab Manual S15 v3Bryan CramptonNo ratings yet
- Namma Kalvi 11th Bio-Botany Study Material English Medium 219431Document51 pagesNamma Kalvi 11th Bio-Botany Study Material English Medium 219431Prasanth PrasanthNo ratings yet
- High Yield Biochemistry PDFDocument41 pagesHigh Yield Biochemistry PDFKyle Broflovski100% (2)
- Problem SetDocument69 pagesProblem SetchemggNo ratings yet
- Lesson 4-7: Molecular Structure of DNA, RNA, and Proteins: Rhoda S.R. Cayanan, RPH, LPTDocument75 pagesLesson 4-7: Molecular Structure of DNA, RNA, and Proteins: Rhoda S.R. Cayanan, RPH, LPTKarma Akira100% (1)
- Bioinfo Final PracticalDocument66 pagesBioinfo Final PracticalPolu ChattopadhyayNo ratings yet
- Molecular Basis of InheritanceDocument18 pagesMolecular Basis of Inheritancegajendran bNo ratings yet
- Chemistry Sample ISC Board Project-PolymersDocument16 pagesChemistry Sample ISC Board Project-PolymersCode Lyoko Fan 10No ratings yet
- Chapter 2 ExamKrackers Biomolecules GeneticsDocument8 pagesChapter 2 ExamKrackers Biomolecules GeneticsPatricia CosNo ratings yet
- BIO 362 Exam 2 PackageDocument62 pagesBIO 362 Exam 2 PackageNerdy Notes Inc.100% (1)
- Coronaviruses Methods and Protocols - Helena Jane Maier 2015Document299 pagesCoronaviruses Methods and Protocols - Helena Jane Maier 2015Geronimo Sanchez VicarioNo ratings yet
- LaudabletasksDocument25 pagesLaudabletasksiremsenakNo ratings yet
- TUTORIAL: DNA BIOLOGY and TECHNOLOGY 1. Describe The Biochemical CompositionDocument6 pagesTUTORIAL: DNA BIOLOGY and TECHNOLOGY 1. Describe The Biochemical Compositionaesha89No ratings yet
- School - Diagnosis of Genetic DisordersDocument33 pagesSchool - Diagnosis of Genetic DisordersBoobesh Venkata Ramanan100% (1)
- Bio Practice Questions For Final Exam Biol 1101Document5 pagesBio Practice Questions For Final Exam Biol 1101Megadirectioner 21No ratings yet
- BiochemDocument383 pagesBiochemtylermedNo ratings yet
- DNASequencing - Lecture 7 PDFDocument34 pagesDNASequencing - Lecture 7 PDFkasmshahabNo ratings yet
- (Ricardo v. Lloyd) Morphology Methods Cell and Mo (B-Ok - Xyz)Document439 pages(Ricardo v. Lloyd) Morphology Methods Cell and Mo (B-Ok - Xyz)dedi sunarto100% (1)