Professional Documents
Culture Documents
Protn STR
Protn STR
Uploaded by
preethi.elizabeth20220 ratings0% found this document useful (0 votes)
1 views1 pageOriginal Title
protn str
Copyright
© © All Rights Reserved
Available Formats
DOCX, PDF, TXT or read online from Scribd
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
© All Rights Reserved
Available Formats
Download as DOCX, PDF, TXT or read online from Scribd
Download as docx, pdf, or txt
0 ratings0% found this document useful (0 votes)
1 views1 pageProtn STR
Protn STR
Uploaded by
preethi.elizabeth2022Copyright:
© All Rights Reserved
Available Formats
Download as DOCX, PDF, TXT or read online from Scribd
Download as docx, pdf, or txt
You are on page 1of 1
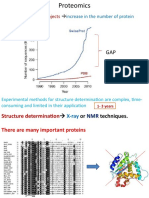
Proteins are essential to life, and understanding their structure can facilitate a
mechanistic understanding of their function. Through an enormous
experimental effort1,2,3,4, the structures of around 100,000 unique proteins
have been determined5, but this represents a small fraction of the billions of
known protein sequences6,7. Structural coverage is bottlenecked by the
months to years of painstaking effort required to determine a single protein
structure. Accurate computational approaches are needed to address this gap
and to enable large-scale structural bioinformatics. Predicting the three-
dimensional structure that a protein will adopt based solely on its amino acid
sequence—the structure prediction component of the ‘protein folding
problem’8—has been an important open research problem for more than
50 years9. Despite recent progress10,11,12,13,14, existing methods fall far short of
atomic accuracy, especially when no homologous structure is available. Here
we provide the first computational method that can regularly predict protein
structures with atomic accuracy even in cases in which no similar structure is
known. We validated an entirely redesigned version of our neural network-
based model, AlphaFold, in the challenging 14th Critical Assessment of protein
Structure Prediction (CASP14)15, demonstrating accuracy competitive with
experimental structures in a majority of cases and greatly outperforming other
methods. Underpinning the latest version of AlphaFold is a novel machine
learning approach that incorporates physical and biological knowledge about
protein structure, leveraging multi-sequence alignments, into the design of the
deep learning algorithm.
You might also like
- Tramontano A. - Protein Structure Prediction 2007 - t1v3Document46 pagesTramontano A. - Protein Structure Prediction 2007 - t1v3Carlos LopeNo ratings yet
- The Role of Protein Structure in Genomics: MinireviewDocument5 pagesThe Role of Protein Structure in Genomics: MinireviewAakriti SharmaNo ratings yet
- Protein Structure Determination and Prediction A Review of TechniquesDocument14 pagesProtein Structure Determination and Prediction A Review of TechniquesIJRRRNo ratings yet
- AlphaFold - Laterst - s41586 021 03819 2Document12 pagesAlphaFold - Laterst - s41586 021 03819 2pafili2762No ratings yet
- Highly Accurate Protein Structure Prediction With Alphafold: ArticleDocument12 pagesHighly Accurate Protein Structure Prediction With Alphafold: ArticleSebastian KmiecikNo ratings yet
- Research Papers On Protein FoldingDocument5 pagesResearch Papers On Protein Foldingshvximwhf100% (1)
- Innovative Computing Review (ICR) : Issn: 2791-0024 ISSN: 2791-0032 HomepageDocument17 pagesInnovative Computing Review (ICR) : Issn: 2791-0024 ISSN: 2791-0032 HomepageINNOVATIVE COMPUTING REVIEWNo ratings yet
- HardwareDocument16 pagesHardwarepaulogameiroNo ratings yet
- Poing PDFDocument16 pagesPoing PDFAanNo ratings yet
- Structural GenomicsDocument6 pagesStructural GenomicsRabia KhattakNo ratings yet
- 2 ProteoDocument17 pages2 ProteoPorntip ChaimaneeNo ratings yet
- Introduction To Protein Folding For Physicists: 1 Why Study Proteins?Document53 pagesIntroduction To Protein Folding For Physicists: 1 Why Study Proteins?Alishba Faixan100% (1)
- Task EenDocument6 pagesTask EenImran JavedNo ratings yet
- Article: A de Novo Protein Binding Pair by Computational Design and Directed EvolutionDocument11 pagesArticle: A de Novo Protein Binding Pair by Computational Design and Directed EvolutionSagar KhareNo ratings yet
- Protein Folding and Assembly Literature ReviewDocument6 pagesProtein Folding and Assembly Literature ReviewSamuel Amon OkumuNo ratings yet
- CPPS SandhyaDocument17 pagesCPPS SandhyaKenesei GyörgyNo ratings yet
- Combinatorial Libraries: Overview of Applications in Chemical BiologyDocument10 pagesCombinatorial Libraries: Overview of Applications in Chemical BiologyazzaassNo ratings yet
- In Silico Prediction and Characterization of Protein Post Translational Modifications - 2016 - Journal of ProteomicsDocument11 pagesIn Silico Prediction and Characterization of Protein Post Translational Modifications - 2016 - Journal of ProteomicsmaikruspeNo ratings yet
- 2009 Brief Bioinform 10 378-391Document14 pages2009 Brief Bioinform 10 378-391mbrylinskiNo ratings yet
- Struct GenomDocument7 pagesStruct GenomNaidelyn HernandezNo ratings yet
- On The Accuracy of Homology Modeling and Sequence AlignmentDocument10 pagesOn The Accuracy of Homology Modeling and Sequence AlignmentCRISTIAN GABRIEL ZAMBRANO VEGANo ratings yet
- Alex - S Protein Folding For SCIENCE EPQ StuffDocument3 pagesAlex - S Protein Folding For SCIENCE EPQ StuffFunkymaleNo ratings yet
- Protein Folding ThesisDocument4 pagesProtein Folding ThesisBuyEssaysAnchorage100% (2)
- Babbitt PDFDocument2 pagesBabbitt PDFSamra KanwalNo ratings yet
- Papers Abstract To ReadDocument2 pagesPapers Abstract To ReadSapan MandloiNo ratings yet
- SESNetDocument19 pagesSESNetThales WillianNo ratings yet
- Protein Folding Research PaperDocument5 pagesProtein Folding Research Paperqhujvirhf100% (1)
- In Silico Attributes of Cold Resistance Protein 1 (Crp1) F Rom Brassica OleraceaeDocument8 pagesIn Silico Attributes of Cold Resistance Protein 1 (Crp1) F Rom Brassica OleraceaeresearchinventyNo ratings yet
- Mol - Modelling of BCL-2 FinalDocument37 pagesMol - Modelling of BCL-2 FinalVannakumar SubramanianNo ratings yet
- Searching For Folded Proteins And: in Vitro in SilicoDocument8 pagesSearching For Folded Proteins And: in Vitro in SilicoVenkata Suryanarayana GorleNo ratings yet
- ZippingDocument21 pagesZippingzaid azharNo ratings yet
- 2015 Article 14 Twilight ZoneDocument11 pages2015 Article 14 Twilight Zoneadiav PakNo ratings yet
- Bio in For Ma TicsDocument2 pagesBio in For Ma TicsKrystal PhillipNo ratings yet
- De Novo Proteins With Life-Sustaining Functions Are Structurally DynamicDocument1 pageDe Novo Proteins With Life-Sustaining Functions Are Structurally DynamicEloisa Marie IglesiasNo ratings yet
- Introducing Protein Folding Using Simple ModelsDocument32 pagesIntroducing Protein Folding Using Simple Modelstestonly261No ratings yet
- Experimental Investigation of Protein Folding and MisfoldingDocument11 pagesExperimental Investigation of Protein Folding and MisfoldingJean Pierre Chastre LuzaNo ratings yet
- Basics of ProteomicsDocument7 pagesBasics of ProteomicskonndaaiahNo ratings yet
- Intrinsically Unstructured Proteins: Re-Assessing The Protein Structure-Function ParadigmDocument11 pagesIntrinsically Unstructured Proteins: Re-Assessing The Protein Structure-Function ParadigmJose DanielNo ratings yet
- (Eisenberg@@) (2000) (NA) (X) Protein Function PDFDocument4 pages(Eisenberg@@) (2000) (NA) (X) Protein Function PDFbyominNo ratings yet
- Functional Genomics and Proteomics As A Foundation For Systems BiologyDocument10 pagesFunctional Genomics and Proteomics As A Foundation For Systems BiologyNidhi JaisNo ratings yet
- Bioinformatics TM6Document30 pagesBioinformatics TM6subhanilchakrabortyNo ratings yet
- Algorithms and Methods in Structural Bioinformatics (Computational Biology) (Nurit Haspel (Editor) Etc.)Document120 pagesAlgorithms and Methods in Structural Bioinformatics (Computational Biology) (Nurit Haspel (Editor) Etc.)Sathish KumarNo ratings yet
- Protein Structure Prediction Using Homology ModelingDocument11 pagesProtein Structure Prediction Using Homology Modelingthelmamapangela64No ratings yet
- Intro To Protein FoldingDocument53 pagesIntro To Protein FoldingRickey Castro100% (1)
- Todd O. Yeates, Todd S. Norcross and Neil P. King - Knotted and Topologically Complex Proteins As Models For Studying Folding and StabilityDocument14 pagesTodd O. Yeates, Todd S. Norcross and Neil P. King - Knotted and Topologically Complex Proteins As Models For Studying Folding and StabilityLokosooNo ratings yet
- Proteomics Tools and Applications: A. EinsteinDocument62 pagesProteomics Tools and Applications: A. EinsteintejalNo ratings yet
- 2010 Proteins 78 118-134Document17 pages2010 Proteins 78 118-134mbrylinskiNo ratings yet
- From Words To Literature in Structural Proteomics: InsightDocument10 pagesFrom Words To Literature in Structural Proteomics: Insightcomputer_genius360100% (1)
- Unit 1: Structural GenomicsDocument4 pagesUnit 1: Structural GenomicsLavanya ReddyNo ratings yet
- Joost Snijder: Extending The Boundaries of Native Mass Spectrometry To Study Virus Structure and AssemblyDocument218 pagesJoost Snijder: Extending The Boundaries of Native Mass Spectrometry To Study Virus Structure and AssemblyVukosava Milic TorresNo ratings yet
- On The Entropy of Protein Families: John P. Barton Arup K. Chakraborty Simona Cocco Hugo Jacquin Rémi MonassonDocument27 pagesOn The Entropy of Protein Families: John P. Barton Arup K. Chakraborty Simona Cocco Hugo Jacquin Rémi MonassonDr. K Lalitha GuruprasadNo ratings yet
- Protein Structure Prediction ThesisDocument8 pagesProtein Structure Prediction ThesisWriteMyThesisPaperManchester100% (2)
- Discordancia OMICSDocument7 pagesDiscordancia OMICSLider_rojo88No ratings yet
- Molecular Dynamics Simulations of Membrane Proteins: Philip C. Biggin and Peter J. BondDocument18 pagesMolecular Dynamics Simulations of Membrane Proteins: Philip C. Biggin and Peter J. BondKübra KahveciNo ratings yet
- Practical Applications of Proteomics-A Technique For Large-Scale Study of Proteins: An OverviewDocument4 pagesPractical Applications of Proteomics-A Technique For Large-Scale Study of Proteins: An OverviewVishwas gargNo ratings yet
- Genome Sequencing Projects: Increase in The Number of Protein SequencesDocument27 pagesGenome Sequencing Projects: Increase in The Number of Protein SequencesBiju ThomasNo ratings yet
- 10.IJAEST Vol No 7 Issue No 2 Human Protein Function Prediction An Overview 239 244Document6 pages10.IJAEST Vol No 7 Issue No 2 Human Protein Function Prediction An Overview 239 244helpdesk9532No ratings yet
- Reviews: Advances in Protein Structure Prediction and DesignDocument17 pagesReviews: Advances in Protein Structure Prediction and DesignvenkatavamsibharadwajNo ratings yet
- Logical Modeling of Biological SystemsFrom EverandLogical Modeling of Biological SystemsLuis Fariñas del CerroNo ratings yet
- High School Biology: Comprehensive Content for Cell & Molecular BiologyFrom EverandHigh School Biology: Comprehensive Content for Cell & Molecular BiologyNo ratings yet