Professional Documents
Culture Documents
Unit-Ii: Chemical Biology
Unit-Ii: Chemical Biology
Uploaded by
Surya Prakash KabiOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Unit-Ii: Chemical Biology
Unit-Ii: Chemical Biology
Uploaded by
Surya Prakash KabiCopyright:
Available Formats
UNIT-II
Chemical biology
Books: Biochemistry, Voet and Voet
Principles of Biochemostry, Lehninger
Enzymes (History)
First discovered by Eduard Buchner in 1897
who observed that yeast extracts can ferment
sugar to alcohol
This proved that fermentation was promoted
by molecules that continued to function when
removed from cells
The first enzyme to be purified and
crystallized was urease in 1926; these
crystals consisted entirely of protein
Later, pepsin, trypsin and other digestive
proteins were isolated and determined to be
purely protein as well
Enzymes are the catalysts of nature.
With the exception of catalytic RNA, all enzymes are
proteins.
Catalyst alter the rate of a chemical reaction without
undergoing a permanent change in structure.
Catalytic activity is dependant upon native
conformation; if it is lost, then catalytic activity is
lost as well
All levels of protein architecture must be intact and
correct for enzymes to perform their functions
They range in molecular weights from 12,000 to over
1 million
Enzymes
Simple Enzymes: composed of whole proteins
Complex Enzymes: composed of protein plus a relatively small
organic molecule
holoenzyme = apoenzyme + prosthetic group / coenzyme
A prosthetic group describes a small organic or metalloorganic molecule
bound to the apoenzyme by covalent bonds.
When the binding between the apoenzyme and non-protein
components is non-covalent, the small organic molecule is called a
coenzyme.
Coenzymes serve as transient carriers of specific functional groups.
They often come from vitamins (organic nutrients required in small amounts in the
diet)
Enzyme Classification
Oxidoreductases Act on many chemical groupings to add or remove
hydrogen atoms (transfer of electrons).
Transferases Transfer functional groups between donor and
acceptor molecules. Kinases are specialized
transferases that regulate metabolism by
transferring phosphate from ATP to other
molecules.
Hydrolases Add water across a bond, hydrolyzing it.
Lyases Add water, ammonia or carbon dioxide across
double bonds, or remove these elements to
produce double bonds.
Isomerases Carry out many kinds of isomerization: L to D
isomerizations, mutase reactions (shifts of
chemical groups) and others.
Ligases Catalyze reactions in which two chemical groups
are joined (or ligated) with the use of energy from
ATP.
International Classification of Enzyme
How enzymes work
Enzymes provide specific environments in which
chemical reactions that dont normally proceed under
neutral pH, mild temperature, and aqueous
environment conditions can occur
This region is a pocket on the enzyme known as the
active site
The molecule that is bound to the active site and
acted upon by the enzyme is called the substrate
The two together form what is known as the enzyme-
substrate complex
The function of an enzyme catalyst is to increase the
rate of a chemical reaction, not affect is equilibrium
Therefore, enzymes dont make more product, they
just make product faster
Active Site
The area of an enzyme that binds to the substrate
Structure has a unique geometric shape that is designed to
fit the molecular shape of the substrate
Each enzyme is substrate specific
Thus the active site that is complementary to the geometric
shape of a substrate molecule
Active site is lined with residues and sometimes contains a co-
factor
Active site residues have several important properties:
Charge [partial, dipoles, helix dipole]
pKa
Hydrophobicity
Flexibility
Reactivity
Substrate Binding specificity
Complementarity
Geometric
Electronic (electrostatic)
Stereospecificity (enzymes and
substrates are chiral)
1. Lock and Key model
2. Induced Fit model
Enzyme active site
Chymotrypsin (Cuts next to Hydrophobic Groups)
Trypsin (Cuts next to Arg & Lys)
Elastase (Cuts next to Val & Thr)
An enzyme binds a substrate in a region called the active site
Only certain substrates can fit the active site
Amino acid R groups in the active site help substrate bind
Lock and Key Model
Enzyme structure flexible, not rigid
Enzyme and active site adjust shape to bind substrate
Increases range of substrate specificity
Shape changes also improve catalysis during reaction
- transition-state like configuration
Induced Fit Model
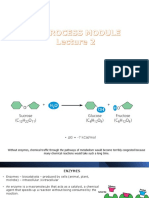
Enzyme-Substrate Interaction
Sucrose Glucose and Fructose
sucrase
hydrolysis
Hexokinase undergoes a conformational change upon
binding to a substrate
red: before substrate-binding
green: after substrate-binding
Effect of Temperature
Effect of pH
The pH-rate profile of an enzyme is a function of the
pK
a
values of the catalytic groups in the enzyme
a group is
catalytically
active in its basic
form
a group is
catalytically
active in its acidic
form
Dependence of lysozyme activity on the pH of the reaction
Asp 52
Glu 35
The pH at which
enzyme is 50% active
Carboxypeptidase A catalyzes the hydrolysis of the
C-terminal peptide
You might also like
- Service Manual (AT) - J Transmission (AT)Document16 pagesService Manual (AT) - J Transmission (AT)Николай КирилловNo ratings yet
- TP0000065 (E) Platinum RTDs PDFDocument17 pagesTP0000065 (E) Platinum RTDs PDFCarlosNo ratings yet
- Maximo in Oil and GasDocument29 pagesMaximo in Oil and GasDia Putranto Harmay100% (1)
- Cosmopolitan - May 2020 USA PDFDocument134 pagesCosmopolitan - May 2020 USA PDFmagazin email100% (4)
- Materi 2Document34 pagesMateri 2siti purnamaNo ratings yet
- Title: Lecture (9) Enzymes Present By: Associate Prof - Ingy BadawyDocument58 pagesTitle: Lecture (9) Enzymes Present By: Associate Prof - Ingy BadawyAbdo ElsaidNo ratings yet
- Enzymes PPTDocument40 pagesEnzymes PPTJaisy PatelNo ratings yet
- Enzyme and Enzyme KineticsDocument61 pagesEnzyme and Enzyme KineticsNaiomiNo ratings yet
- B Topic 7 - Enzymes - Structure, Functions, and ClassificationDocument34 pagesB Topic 7 - Enzymes - Structure, Functions, and Classificationfantasticedwin254No ratings yet
- "Helper" Protein Molecules: EnzymesDocument7 pages"Helper" Protein Molecules: EnzymesMuhammad Junaid IqbalNo ratings yet
- SCE3204 Lecture 2Document31 pagesSCE3204 Lecture 2ainomugisha arnoldNo ratings yet
- Enzymes and Vitamins CompressedDocument14 pagesEnzymes and Vitamins CompressedfredkaseydiNo ratings yet
- Enzymes PresDocument57 pagesEnzymes Pres4tqpxmts7zNo ratings yet
- Enzymes: by Nura Malahayati THP-FP UnsriDocument71 pagesEnzymes: by Nura Malahayati THP-FP UnsriIkhsan muttaqinNo ratings yet
- Enzymes - BiochemistryDocument40 pagesEnzymes - Biochemistrysunil patelNo ratings yet
- Babi Pandey Biochemistry Vii Semester: Submitted To: Anil RegmiDocument38 pagesBabi Pandey Biochemistry Vii Semester: Submitted To: Anil RegmiBabi PandeyNo ratings yet
- EnzymesDocument99 pagesEnzymesSamuel AgulueNo ratings yet
- Enzymes PPTDocument39 pagesEnzymes PPTsunil patelNo ratings yet
- EnzymesDocument65 pagesEnzymesnaghalfatih100% (2)
- EnzymesDocument53 pagesEnzymesMayuri DhanushwarNo ratings yet
- Enzymes 1Document54 pagesEnzymes 1Mayuri DhanushwarNo ratings yet
- Protein in Human BodyDocument47 pagesProtein in Human BodyYulia KasihNo ratings yet
- Enzymes:: The Nature's CatalystsDocument51 pagesEnzymes:: The Nature's Catalystskrk100% (1)
- Unit VIII - Enzymes - MCONDocument30 pagesUnit VIII - Enzymes - MCONikram ullah khanNo ratings yet
- EnzymologyDocument47 pagesEnzymologyTererai Lalelani Masikati Hove100% (1)
- BCH 209: Introductory Enzymology: Lecturer: Dr. O. J. AvwiorokoDocument35 pagesBCH 209: Introductory Enzymology: Lecturer: Dr. O. J. Avwiorokobuhari rabiuNo ratings yet
- Enzyme: Ayesha Shafi Pharm-D, (P.U.), M. Phil. Pharmaceutical Chemistry (P.U.)Document34 pagesEnzyme: Ayesha Shafi Pharm-D, (P.U.), M. Phil. Pharmaceutical Chemistry (P.U.)Shafaqat Ghani Shafaqat Ghani100% (2)
- Enzymes PresDocument34 pagesEnzymes PresRasmia SalamNo ratings yet
- EnzymeDocument192 pagesEnzymeKishoreNo ratings yet
- EnzymesDocument41 pagesEnzymesaa9140697874No ratings yet
- 1FFF11B1BD16627EE05400144FEB5F70.pptDocument63 pages1FFF11B1BD16627EE05400144FEB5F70.pptNur AishaNo ratings yet
- Biochem 1 - NOMENCLATURE OF ENZYMESDocument51 pagesBiochem 1 - NOMENCLATURE OF ENZYMESNyasha Peter Manamike100% (1)
- Enzymes Dr. Madhur GuptaDocument89 pagesEnzymes Dr. Madhur GuptaAarohiNo ratings yet
- BIOCHEMISTRY LECTURE NotesDocument31 pagesBIOCHEMISTRY LECTURE NotesFREDRICK OUNDONo ratings yet
- Enzymes ClassDocument35 pagesEnzymes ClassVivek kumarNo ratings yet
- EnzymesDocument5 pagesEnzymeseyosiyas tekleweyinNo ratings yet
- Enzymes Part IIDocument51 pagesEnzymes Part IIjasonenmanuel10No ratings yet
- EnzymesDocument46 pagesEnzymesHighlifeNo ratings yet
- Biochemistry - Enzyme-Protein Function and Metabolism 20180202Document78 pagesBiochemistry - Enzyme-Protein Function and Metabolism 20180202Artika Muliany TindaonNo ratings yet
- Enzyme 1Document115 pagesEnzyme 1fikaduNo ratings yet
- Động Hoá Học Duoc-32-112Document81 pagesĐộng Hoá Học Duoc-32-112Nguyễn Đình TrườngNo ratings yet
- BMS131 Lec05Document19 pagesBMS131 Lec05mariam tarekNo ratings yet
- Enzyme Structure, Classification and Mechanism of ActionDocument37 pagesEnzyme Structure, Classification and Mechanism of ActionYudha Rizki WahyudiNo ratings yet
- Classification of Enzymes: Sadia SayedDocument25 pagesClassification of Enzymes: Sadia SayedmiadelfiorNo ratings yet
- 1 Enzyme Regulation in Biochemical PathwaysDocument29 pages1 Enzyme Regulation in Biochemical PathwayshadeelNo ratings yet
- CHEM 205 LECTURE 4 Enzyme KineticsDocument47 pagesCHEM 205 LECTURE 4 Enzyme KineticsHannah BuquironNo ratings yet
- EnzymesDocument40 pagesEnzymesAmal100% (2)
- 3.5 Enzymes 2Document64 pages3.5 Enzymes 2Alondra SagarioNo ratings yet
- EnzymesDocument36 pagesEnzymesAyush GoelNo ratings yet
- Lecture 2Document18 pagesLecture 2warden tambiNo ratings yet
- Enzyme IntroductionDocument85 pagesEnzyme IntroductionChemistry DepartmentNo ratings yet
- Biomolecules and Cells:: Mr. Derrick Banda MSC, BSCDocument69 pagesBiomolecules and Cells:: Mr. Derrick Banda MSC, BSCAmon Sangulube100% (1)
- Biochem NotesDocument52 pagesBiochem NotesAnonymous 4Ib1ByFC100% (2)
- Btech Great Intro To Enzymes For MLB 104 UseDocument75 pagesBtech Great Intro To Enzymes For MLB 104 Usemaxwell amponsahNo ratings yet
- Chapter 4 Metabolism and Metabolic DisordersDocument77 pagesChapter 4 Metabolism and Metabolic DisordersCumar MaxamuudNo ratings yet
- Enzymes NPDocument25 pagesEnzymes NPBharatShethNo ratings yet
- Clinical Chemistry 2 First GradingDocument25 pagesClinical Chemistry 2 First GradingMHEKAELLA SAMSONNo ratings yet
- Biochemistry 1: - Biochemistry of Amino Acids - Biochemistry of Proteins - Portrait of Allosteric ProteinDocument55 pagesBiochemistry 1: - Biochemistry of Amino Acids - Biochemistry of Proteins - Portrait of Allosteric ProteinHiba N IkhmyesNo ratings yet
- Lesson 1 Enzyme Action PowerpointDocument18 pagesLesson 1 Enzyme Action Powerpointtbrook2017No ratings yet
- EnzymesDocument71 pagesEnzymesSaran DNo ratings yet
- Enzyme StructureDocument2 pagesEnzyme StructurePrince OycoNo ratings yet
- Bio Lec 9 Enzyme 1Document41 pagesBio Lec 9 Enzyme 1hadeelabouriNo ratings yet
- R11. Enzymes and Other Proteins: Henedina A. Maini, RPH LecturerDocument33 pagesR11. Enzymes and Other Proteins: Henedina A. Maini, RPH LecturerChyra OrpillaNo ratings yet
- A-level Biology Revision: Cheeky Revision ShortcutsFrom EverandA-level Biology Revision: Cheeky Revision ShortcutsRating: 5 out of 5 stars5/5 (5)
- Tractor Caterpillar d7r Renr5627 Renr5627 P Esquema HidraulicoDocument32 pagesTractor Caterpillar d7r Renr5627 Renr5627 P Esquema HidraulicoRuberli RJNo ratings yet
- (Studies in Fuzziness and Soft Computing 246) Piedad Brox, Iluminada Baturone, Santiago Sánchez-Solano (Auth.) - Fuzzy logic-Springer-VerlagDocument182 pages(Studies in Fuzziness and Soft Computing 246) Piedad Brox, Iluminada Baturone, Santiago Sánchez-Solano (Auth.) - Fuzzy logic-Springer-VerlagRXNo ratings yet
- 301 Signal & System Analysis SolDocument165 pages301 Signal & System Analysis SolAbelito ReyesNo ratings yet
- ENCh 11Document43 pagesENCh 11ShyamRathiNo ratings yet
- Bip 0071-2014Document130 pagesBip 0071-2014anishdev6No ratings yet
- Engineering Mechanics ProblemDocument71 pagesEngineering Mechanics ProblemEu Aumentado0% (1)
- Geologic Processes On EarthDocument38 pagesGeologic Processes On EarthTrisha May FloresNo ratings yet
- Diagnosing and Repairing Carbonation in Concrete Structures PDFDocument7 pagesDiagnosing and Repairing Carbonation in Concrete Structures PDFflucayNo ratings yet
- Cobalt DriveDocument5 pagesCobalt DriveEifla GonzalesNo ratings yet
- Design of PLC Based Speed Control of DC Motor Using PI ControllerDocument4 pagesDesign of PLC Based Speed Control of DC Motor Using PI ControllerTricia Mae EvangelistaNo ratings yet
- PWS6500 Hardware ManualDocument15 pagesPWS6500 Hardware ManualOmar Alberto Quisbert TapiaNo ratings yet
- Televisión y VideograbadoraDocument71 pagesTelevisión y VideograbadoraMoyses MoyNo ratings yet
- Pengaruh Laju Erosi Terhadap Umur PipaDocument6 pagesPengaruh Laju Erosi Terhadap Umur PipaDede Si Engghe SurenggheNo ratings yet
- STC1100T6单行本 巴西自动底盘-中英文版20231018Document31 pagesSTC1100T6单行本 巴西自动底盘-中英文版20231018Matheus PiresNo ratings yet
- NanoparticlesDocument13 pagesNanoparticlesArslan JigarNo ratings yet
- MPL Job Application Form: That All Your Information Must Be Typed in The Spaces Provided)Document4 pagesMPL Job Application Form: That All Your Information Must Be Typed in The Spaces Provided)Owere patrickNo ratings yet
- Wa0003Document74 pagesWa0003Rohit ThakurNo ratings yet
- Sample Project Report On Financial Analysis at B D K LTD HubaliDocument102 pagesSample Project Report On Financial Analysis at B D K LTD HubalihareeshngvNo ratings yet
- CriteriaDocument7 pagesCriteriaShajahan ShagulNo ratings yet
- Trafoindo Catalogue Oil Immersed Transformers PDFDocument2 pagesTrafoindo Catalogue Oil Immersed Transformers PDFMustavaNo ratings yet
- ASME Section X InterpretationsDocument6 pagesASME Section X InterpretationsAnonymous UoHUagNo ratings yet
- Spare List SPLC PDFDocument7 pagesSpare List SPLC PDFmsmagaNo ratings yet
- GPRS & MMS Settings: IndosatDocument6 pagesGPRS & MMS Settings: IndosatTaufik Acau NovaNo ratings yet
- O&M For Elevator EnglishDocument63 pagesO&M For Elevator Englishdo phuong100% (4)
- GL TablesDocument31 pagesGL TablesGanga KanimozhiNo ratings yet
- Eco NicalDocument13 pagesEco Nicalluizfellipe95No ratings yet