Professional Documents
Culture Documents
Poster Final
Poster Final
Uploaded by
uditmajumder18102002Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Poster Final
Poster Final
Uploaded by
uditmajumder18102002Copyright:
Available Formats
Isolation Of Bacteria And Testing For Gelatinase Enzymatic Activity
~Sagnik Biswas, Dipannita Bhattacharya (presenting author), Suman Chakraborty, Poulami Pal, Udit Majumdar, Sayantan Sarkar, Ananya Baghchi, Subhechchha Majhi, Anwesha Goldar, Somalika Pramanik,
Amina khatun, Hishma Fareah, Nisae A Memon, Ananya Debnath, Raju Mondal, Swastik Maity, Sanjana Guho Neogi.
Semister 1, Department Of Microbiology, Maulana Azad Collage,
8 Rafi Ahmed Kidwai Road, Kolkata, 700013
Objective: Five potential strains with suspected gelatinase activity were pure-cultured, and their gram-nature and morphology were determined.
Their biochemical characterization also carried out with respect to nitrate reductase, catalase and urease activity.
We first collected 5 bacterial colony from collage campus.
We have prepared pure culture of those strains by
streaking. And proceeded through below mention staps as
follows-
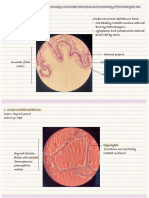
strains Cell shape Colonial colour Gram Colony
shape charecter number
1 spherical filamentous white +ve 14
2 Rod shaped circular orange +ve 9
3 spherical irregular Creamish yellow +ve 6
4 spherical circular Light yellow +ve 4
5 spherical circular Yellowish orange +ve 17
Gelatinase producing bacteria isolation
• Media =>Minimal media +15% gelatin+0.5% agar.
• Stabbing in testube and incubated for 1 week.
• rechecked and confirmed by HgCl2 precipitaton process.
Strain no. Observation- HgCl2 precipitation Gelatinase enzyme
Media +/- Gelatinase? Interesting!!!!
• Cancer therapy-exhibit proteolytic activity against
1 solid no - tumour associated ECM proteins,potentially aiding in
tumour invasion and metastasis.
2 liquified yes + • Bioremedian application-degrades
microplastics,hydrocarbons,dyes,pesticides.
3 solid no -
• Wound healing-can degrade collagen,a major
component of extra cellular matrix(ECM) facilitating
4 solid no - tissue remodeling and wound healing
• Enzyme engineering-enhancing the activity,specificity
5 liquified yes +
and stability of enzymes.
Chemical tests:
Urease test- strain no. Observation-Change Urease +/-
• Principle-identification of urease producing strains which convert of media colour
urea to ammonia and co2. 2 no -
• Media-Christensen’s urea agar + phenol red
• Method-1.slants using prepared broth are prepared,2.streaking 5 no -
of thick pure culture,3.incubation.
Catalase test- Strain no. Observation-
bubble Catalase +/-
• Principle- detection of preascence of catalase enzyme in formation after
bacteria,by addition of H2O2. adding H2O2
• Media-1.L.B.agar,2.3% H2O2 solution.
• Method-1.a drop of water +inoculum,smear formation,2. 2
no -
1 drop H2O2 added.
5 no -
Strain no. Observation- Observation-
Nitrate reductase test- red colour red colour
after adding after adding
• Principle- Detection of nitrate reductase enzyme producing A and B zinc powder
strains which convert NO3- >NO2->NH3+ OFR N2.
2 no no
• Media-tripticase nitrate broth(ph-7.2)
• Other requirements- sulphanilic acid(A), alpha
napthylamine(B),zinc powder. 5 no no
• Method- 5 drops od A+ 5 drops of B+zinc powder.
Current status- currently working on antibiotic resistance checking by amphicillin, kanamycin,methicillin,tetracycline; gelatin quantitative
analysis;16s rRNA profiling.
You might also like
- Guide To Inspections of Low Acid Canned Food Manufacturers - Part 2Document82 pagesGuide To Inspections of Low Acid Canned Food Manufacturers - Part 2william eduardo mercado navarroNo ratings yet
- Morphological and Physiological Identification of MicrobesDocument6 pagesMorphological and Physiological Identification of MicrobesNajela Leila CastroNo ratings yet
- Routine Stool ExaminationDocument2 pagesRoutine Stool ExaminationarvenemartinNo ratings yet
- Identification of UnknownDocument7 pagesIdentification of Unknownkimmynemil80No ratings yet
- Cytology SlidesDocument28 pagesCytology SlidesYasir AliNo ratings yet
- Histopathologic TechniquesDocument11 pagesHistopathologic TechniquesGail SantanderNo ratings yet
- Histopathologic TechniquesDocument10 pagesHistopathologic TechniquesM C0% (1)
- Unit 2 MicrobiologyDocument47 pagesUnit 2 Microbiologykrishkodal0020No ratings yet
- Practical MKS ModuleDocument12 pagesPractical MKS ModuleMohammed HashemNo ratings yet
- Pages From Rajesh Bardale Principles of Forensic Medicine and ToxicologyDocument2 pagesPages From Rajesh Bardale Principles of Forensic Medicine and Toxicologymueen hashmiNo ratings yet
- Microbiology Lab MidTermsDocument2 pagesMicrobiology Lab MidTermsLaurence Bernard Jasper CanlasNo ratings yet
- Micp211 Lab Midterm ReviewerDocument4 pagesMicp211 Lab Midterm ReviewerMAV TAJNo ratings yet
- 14 MycobacteriaDocument3 pages14 MycobacteriaAlthea Jam Grezshyl GaloNo ratings yet
- Lesson5.3 LabDiaofTBDocument3 pagesLesson5.3 LabDiaofTBJAY JR. APONNo ratings yet
- Staining Procedures: 3. Diagnostic Antibody or DNA Probe Mediated StainingDocument2 pagesStaining Procedures: 3. Diagnostic Antibody or DNA Probe Mediated StainingLiah ManlapigNo ratings yet
- IARCCervicalCancerImageBank ColpoMetaDataDocument5 pagesIARCCervicalCancerImageBank ColpoMetaDatasudershannegi7013No ratings yet
- Research Journal of Pharmacognosy and Phytochemistry - PID - 2011-3-6-5Document3 pagesResearch Journal of Pharmacognosy and Phytochemistry - PID - 2011-3-6-5Abd EL HakimNo ratings yet
- PRACTICAL 4 MicrobDocument6 pagesPRACTICAL 4 Microbdomo- kunNo ratings yet
- Staining TechniquesDocument2 pagesStaining TechniquesRonalyn UgatNo ratings yet
- Aubf Lab MidtermDocument8 pagesAubf Lab MidtermNiezel EncalladoNo ratings yet
- Bacte Comprehensive ReviewDocument116 pagesBacte Comprehensive ReviewFaith Theresa OroscoNo ratings yet
- Examination of Fecal and Cloacal Swab Sample ofDocument26 pagesExamination of Fecal and Cloacal Swab Sample ofSapana YadavNo ratings yet
- Pharmacodynamics - ReviewerDocument10 pagesPharmacodynamics - ReviewerGabrielle Marie GonzagaNo ratings yet
- Lecture-4Document9 pagesLecture-4Emmanuel Tristan OlarteNo ratings yet
- Pharmacological and Toxicol Ogical Screening: G o o D M o RN in G !Document43 pagesPharmacological and Toxicol Ogical Screening: G o o D M o RN in G !Saloni MehtaNo ratings yet
- Biochemical Tests BacteriaDocument30 pagesBiochemical Tests BacteriaScribdTranslationsNo ratings yet
- Bacte ReviewerDocument4 pagesBacte ReviewerMaryangel RivalesNo ratings yet
- Stains NileDocument37 pagesStains NileNUBWA MEDUGUNo ratings yet
- Fundamentals of AUBFDocument12 pagesFundamentals of AUBFWho KnowsNo ratings yet
- DIG High Prime DNA Labeling and Detection Starter Kit I: Instruction ManualDocument28 pagesDIG High Prime DNA Labeling and Detection Starter Kit I: Instruction ManualNur SyamimiNo ratings yet
- PCR and Electrophoresis HandoutDocument4 pagesPCR and Electrophoresis HandoutJJJJJJJ123No ratings yet
- 27ab Lipids&Membranes PDFDocument31 pages27ab Lipids&Membranes PDFAkram ZayedNo ratings yet
- Case Analysis DehydrationDocument6 pagesCase Analysis DehydrationDayan CabrigaNo ratings yet
- CeuticsDocument28 pagesCeuticsلوى كمالNo ratings yet
- HTMLE RecallsDocument3 pagesHTMLE Recallsninaalteres123No ratings yet
- Parasitology Short Quiz 1.2Document2 pagesParasitology Short Quiz 1.2JAIRISH YZABELLE SALVADORNo ratings yet
- Physiology Rapid RevisionDocument41 pagesPhysiology Rapid RevisionAsif MohammedNo ratings yet
- Method of EvaluationDocument38 pagesMethod of EvaluationDwyane DickisonNo ratings yet
- Methods of Identification: MethodologiesDocument12 pagesMethods of Identification: MethodologiesNiña Georgette ViñaNo ratings yet
- Cell Proliferation Kit I (MTT) : Cat. No. 11 465 007 001Document4 pagesCell Proliferation Kit I (MTT) : Cat. No. 11 465 007 001AyuAnatriera100% (1)
- Practical No 3. StaphylococciDocument3 pagesPractical No 3. StaphylococciAsyeon GhaziNo ratings yet
- Lab Prep 4 5 PDFDocument3 pagesLab Prep 4 5 PDFGNCDWNo ratings yet
- Using Drosophila For Drug ScreeningDocument15 pagesUsing Drosophila For Drug ScreeningalessandraNo ratings yet
- Lecture4 HowCellsGrow Ch6Document74 pagesLecture4 HowCellsGrow Ch6viraj patilNo ratings yet
- 1 - Practical 1 Histology Introduction To HistologyDocument61 pages1 - Practical 1 Histology Introduction To HistologyAMIRA HELAYELNo ratings yet
- Lecture 4-Prep For Microscopic ExamDocument15 pagesLecture 4-Prep For Microscopic ExamAndreah BaylonNo ratings yet
- Rapid Composting Microbes Used by AICDocument30 pagesRapid Composting Microbes Used by AICGene Gregorio100% (1)
- Virulence FactorsDocument49 pagesVirulence FactorsHira PanhwerNo ratings yet
- Staining Techniques in Microbiology: Simple-Stain-Bacterial-CultureDocument8 pagesStaining Techniques in Microbiology: Simple-Stain-Bacterial-CulturexXjmir142 rocksXxNo ratings yet
- Review: What Three Types of Hemolysis Can Be Observed On A BAP Plate?Document5 pagesReview: What Three Types of Hemolysis Can Be Observed On A BAP Plate?Jonille EchevarriaNo ratings yet
- MIBO3510L Final Practical Review: Feng Long 04-21-2016Document32 pagesMIBO3510L Final Practical Review: Feng Long 04-21-2016Katie MarieNo ratings yet
- Stains & Staining: Faculty: Dr. Rakesh ShardaDocument34 pagesStains & Staining: Faculty: Dr. Rakesh ShardaQueen Lycka P. Serioza100% (1)
- Comparative Study of Dye Degradation by Using Green Synthesized ZnO Nanoparticle and BacillusDocument10 pagesComparative Study of Dye Degradation by Using Green Synthesized ZnO Nanoparticle and BacillusIJRASETPublicationsNo ratings yet
- ReportDocument7 pagesReportWareef AhmadNo ratings yet
- StainDocument5 pagesStainEric RyanNo ratings yet
- B.Tech. IV (CH), Semester - Vii L T P C ES-1: 3 0 0 3 (06 Hours) (02 Hours) (25 Hours)Document17 pagesB.Tech. IV (CH), Semester - Vii L T P C ES-1: 3 0 0 3 (06 Hours) (02 Hours) (25 Hours)ajitsinghrathoreNo ratings yet
- BACTE MODULE 4 Methods of Studying BacteriaDocument5 pagesBACTE MODULE 4 Methods of Studying BacteriaKaylaNo ratings yet
- Yu Hui PDDocument21 pagesYu Hui PDYu HuiNo ratings yet
- Lab 3 - Smear Preparation Simple Staining and Gram StainingDocument29 pagesLab 3 - Smear Preparation Simple Staining and Gram StainingAyman ElkenawyNo ratings yet
- 2 Fixation, Decalcification & DehydrationDocument3 pages2 Fixation, Decalcification & DehydrationNur-Reza MohammadNo ratings yet
- 2022 BSN Exercise No 16 Enzymatic BrowningDocument4 pages2022 BSN Exercise No 16 Enzymatic BrowningElma Kit Aquino GolocanNo ratings yet
- Medicamentos Con Sus PresentacionDocument10 pagesMedicamentos Con Sus Presentacionaisbeli labarcaNo ratings yet
- First/Second Semester B.E.Degree Examination Engineering ChemistryDocument2 pagesFirst/Second Semester B.E.Degree Examination Engineering ChemistryZander IndiaNo ratings yet
- Central Dogma WorksheetDocument8 pagesCentral Dogma WorksheetZyrus AntalanNo ratings yet
- Pilot Operated NRVDocument1 pagePilot Operated NRVBiswanath LenkaNo ratings yet
- Derive The Gibbs-Duhem Equation and Give Its Applications?Document4 pagesDerive The Gibbs-Duhem Equation and Give Its Applications?kannan2030No ratings yet
- 110 The Evaluation of Frying Oils With The P-Anisidine ValueDocument3 pages110 The Evaluation of Frying Oils With The P-Anisidine ValueNicolas AvendañoNo ratings yet
- 2003-Pint - Optimization of Reactive-Element Additions To Improve Oxidation Performance of Alumina-Forming AlloysDocument10 pages2003-Pint - Optimization of Reactive-Element Additions To Improve Oxidation Performance of Alumina-Forming AlloysDavid Hernández EscobarNo ratings yet
- Cosmetics 09 00063 v2Document44 pagesCosmetics 09 00063 v2maizhafiraNo ratings yet
- Cement CoatingsFinalDocument74 pagesCement CoatingsFinalAPEX SON100% (2)
- 6-2 Section ReviewDocument4 pages6-2 Section ReviewStevenNo ratings yet
- Jominy Hardenability Test: ObjectiveDocument1 pageJominy Hardenability Test: ObjectiveAhmadNo ratings yet
- External Corrosion - Introduction To ChemistryDocument93 pagesExternal Corrosion - Introduction To Chemistryzvi cohenNo ratings yet
- Design of Experiment For Optimization of Nitrophenol Reduction by Green Synthesized Silver NanocatalystDocument11 pagesDesign of Experiment For Optimization of Nitrophenol Reduction by Green Synthesized Silver NanocatalystSHERLY KIMBERLY RAMOS JESUSNo ratings yet
- BiosurfactatsDocument6 pagesBiosurfactatsNoor Ul NaeemNo ratings yet
- E 216 Wrap Around Pipe MarkerDocument2 pagesE 216 Wrap Around Pipe MarkerGautam SawhneyNo ratings yet
- Fresh, Mechanical, and Durability Properties of Basalt Fiber-Reinforced Concrete (BFRC) A ReviewDocument17 pagesFresh, Mechanical, and Durability Properties of Basalt Fiber-Reinforced Concrete (BFRC) A ReviewОлег ШибекоNo ratings yet
- 04 Chapter 4Document39 pages04 Chapter 4korangaprakashNo ratings yet
- Good Dispensing PracticesDocument35 pagesGood Dispensing PracticesFaith GabrielNo ratings yet
- 10.1201 b19455 PreviewpdfDocument116 pages10.1201 b19455 PreviewpdfMohammed GhanemNo ratings yet
- Nanocomposites Materials Definitions Typ 767a08f9Document7 pagesNanocomposites Materials Definitions Typ 767a08f9Ahmed TantawyNo ratings yet
- Chemical Periodontal TherapyDocument23 pagesChemical Periodontal Therapyneji_murniNo ratings yet
- Development of Bioprocesses For The Conservation, Detoxification and Value-Addition of Coffee Pulp andDocument2 pagesDevelopment of Bioprocesses For The Conservation, Detoxification and Value-Addition of Coffee Pulp andKatherine Rocio Obeid ManjarrezNo ratings yet
- Catalysis Today: Jangam Ashok, Subhasis Pati, Plaifa Hongmanorom, Zhang Tianxi, Chen Junmei, Sibudjing Kawi TDocument19 pagesCatalysis Today: Jangam Ashok, Subhasis Pati, Plaifa Hongmanorom, Zhang Tianxi, Chen Junmei, Sibudjing Kawi Tfarah al-sudaniNo ratings yet
- Some Basic Concepts of Chemistry 01 - Class Notes - Uday 2025Document38 pagesSome Basic Concepts of Chemistry 01 - Class Notes - Uday 2025BhavishyarohillaNo ratings yet
- 11 Chemistry Notes Ch08 Redox ReactionDocument5 pages11 Chemistry Notes Ch08 Redox ReactionAdarsh GautamNo ratings yet
- Carbon EquivalentDocument13 pagesCarbon Equivalentrasnowmah2012No ratings yet
- TurboVap LV Users ManualDocument48 pagesTurboVap LV Users ManualAhmad HamdounNo ratings yet
- Rouge MonitoringDocument45 pagesRouge Monitoringqa asimasNo ratings yet